Ammar Ahmed Pallikonda Latheef
Department of Computer Science, Johns Hopkins University
Emergent Language Symbolic Autoencoder (ELSA) with Weak Supervision to Model Hierarchical Brain Networks
Apr 15, 2024Abstract:Brain networks display a hierarchical organization, a complexity that poses a challenge for existing deep learning models, often structured as flat classifiers, leading to difficulties in interpretability and the 'black box' issue. To bridge this gap, we propose a novel architecture: a symbolic autoencoder informed by weak supervision and an Emergent Language (EL) framework. This model moves beyond traditional flat classifiers by producing hierarchical clusters and corresponding imagery, subsequently represented through symbolic sentences to improve the clinical interpretability of hierarchically organized data such as intrinsic brain networks, which can be characterized using resting-state fMRI images. Our innovation includes a generalized hierarchical loss function designed to ensure that both sentences and images accurately reflect the hierarchical structure of functional brain networks. This enables us to model functional brain networks from a broader perspective down to more granular details. Furthermore, we introduce a quantitative method to assess the hierarchical consistency of these symbolic representations. Our qualitative analyses show that our model successfully generates hierarchically organized, clinically interpretable images, a finding supported by our quantitative evaluations. We find that our best performing loss function leads to a hierarchical consistency of over 97% when identifying images corresponding to brain networks. This approach not only advances the interpretability of deep learning models in neuroimaging analysis but also represents a significant step towards modeling the intricate hierarchical nature of brain networks.
Deep Labeling of fMRI Brain Networks
May 05, 2023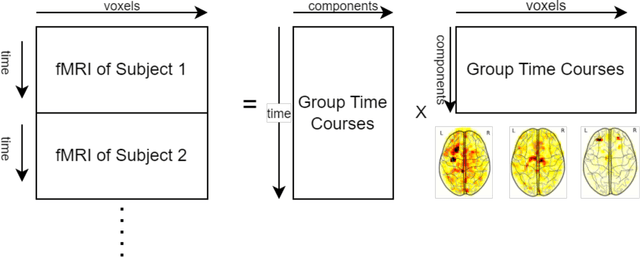
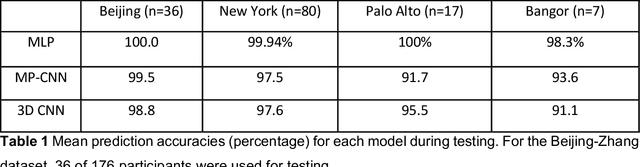
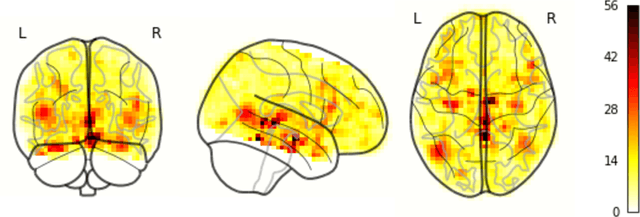
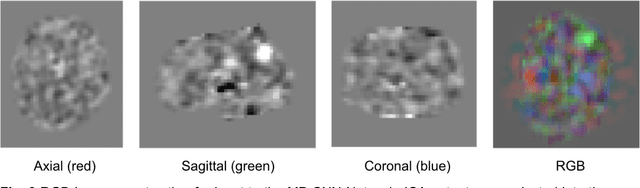
Abstract:Resting State Networks (RSNs) of the brain extracted from Resting State functional Magnetic Resonance Imaging (RS-fMRI) are used in the pre-surgical planning to guide the neurosurgeon. This is difficult, though, as expert knowledge is required to label each of the RSNs. There is a lack of efficient and standardized methods to be used in clinical workflows. Additionally, these methods need to be generalizable since the method needs to work well regardless of the acquisition technique. We propose an accurate, fast, and lightweight deep learning approach to label RSNs. Group Independent Component Analysis (ICA) was used to extract large scale functional connectivity patterns in the cohort and dual regression was used to back project them on individual subject RSNs. We compare a Multi-Layer Perceptron (MLP) based method with 2D and 3D Convolutional Neural Networks (CNNs) and find that the MLP is faster and more accurate. The MLP method performs as good or better than other works despite its compact size. We prove the generalizability of our method by showing that the MLP performs at 100% accuracy in the holdout dataset and 98.3% accuracy in three other sites' fMRI acquisitions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge