Noel C. F. Codella
Data Scaling Laws for Radiology Foundation Models
Sep 16, 2025Abstract:Foundation vision encoders such as CLIP and DINOv2, trained on web-scale data, exhibit strong transfer performance across tasks and datasets. However, medical imaging foundation models remain constrained by smaller datasets, limiting our understanding of how data scale and pretraining paradigms affect performance in this setting. In this work, we systematically study continual pretraining of two vision encoders, MedImageInsight (MI2) and RAD-DINO representing the two major encoder paradigms CLIP and DINOv2, on up to 3.5M chest x-rays from a single institution, holding compute and evaluation protocols constant. We evaluate on classification (radiology findings, lines and tubes), segmentation (lines and tubes), and radiology report generation. While prior work has primarily focused on tasks related to radiology findings, we include lines and tubes tasks to counterbalance this bias and evaluate a model's ability to extract features that preserve continuity along elongated structures. Our experiments show that MI2 scales more effectively for finding-related tasks, while RAD-DINO is stronger on tube-related tasks. Surprisingly, continually pretraining MI2 with both reports and structured labels using UniCL improves performance, underscoring the value of structured supervision at scale. We further show that for some tasks, as few as 30k in-domain samples are sufficient to surpass open-weights foundation models. These results highlight the utility of center-specific continual pretraining, enabling medical institutions to derive significant performance gains by utilizing in-domain data.
CancerGUIDE: Cancer Guideline Understanding via Internal Disagreement Estimation
Sep 09, 2025
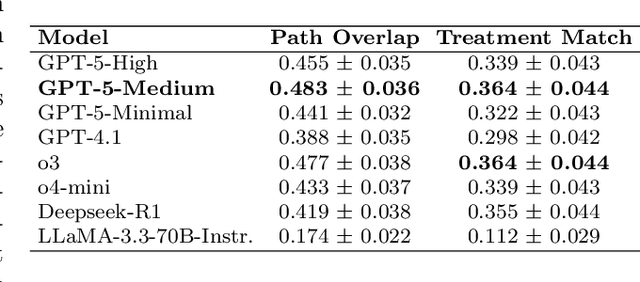
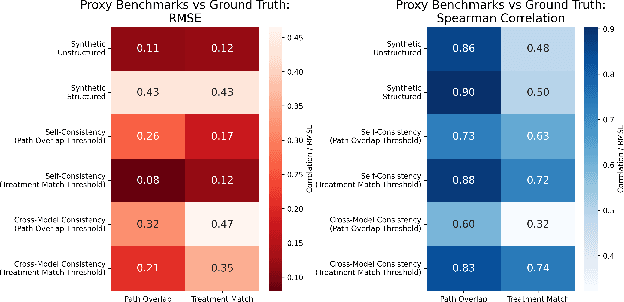
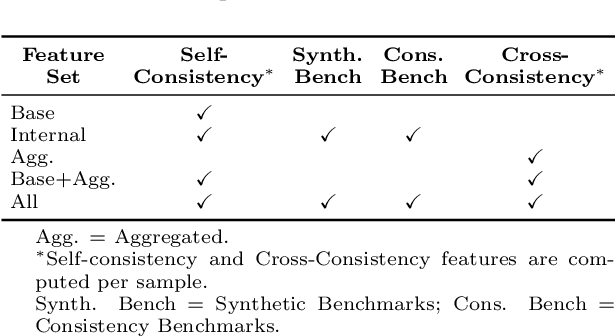
Abstract:The National Comprehensive Cancer Network (NCCN) provides evidence-based guidelines for cancer treatment. Translating complex patient presentations into guideline-compliant treatment recommendations is time-intensive, requires specialized expertise, and is prone to error. Advances in large language model (LLM) capabilities promise to reduce the time required to generate treatment recommendations and improve accuracy. We present an LLM agent-based approach to automatically generate guideline-concordant treatment trajectories for patients with non-small cell lung cancer (NSCLC). Our contributions are threefold. First, we construct a novel longitudinal dataset of 121 cases of NSCLC patients that includes clinical encounters, diagnostic results, and medical histories, each expertly annotated with the corresponding NCCN guideline trajectories by board-certified oncologists. Second, we demonstrate that existing LLMs possess domain-specific knowledge that enables high-quality proxy benchmark generation for both model development and evaluation, achieving strong correlation (Spearman coefficient r=0.88, RMSE = 0.08) with expert-annotated benchmarks. Third, we develop a hybrid approach combining expensive human annotations with model consistency information to create both the agent framework that predicts the relevant guidelines for a patient, as well as a meta-classifier that verifies prediction accuracy with calibrated confidence scores for treatment recommendations (AUROC=0.800), a critical capability for communicating the accuracy of outputs, custom-tailoring tradeoffs in performance, and supporting regulatory compliance. This work establishes a framework for clinically viable LLM-based guideline adherence systems that balance accuracy, interpretability, and regulatory requirements while reducing annotation costs, providing a scalable pathway toward automated clinical decision support.
From Embeddings to Accuracy: Comparing Foundation Models for Radiographic Classification
May 16, 2025Abstract:Foundation models, pretrained on extensive datasets, have significantly advanced machine learning by providing robust and transferable embeddings applicable to various domains, including medical imaging diagnostics. This study evaluates the utility of embeddings derived from both general-purpose and medical domain-specific foundation models for training lightweight adapter models in multi-class radiography classification, focusing specifically on tube placement assessment. A dataset comprising 8842 radiographs classified into seven distinct categories was employed to extract embeddings using six foundation models: DenseNet121, BiomedCLIP, Med-Flamingo, MedImageInsight, Rad-DINO, and CXR-Foundation. Adapter models were subsequently trained using classical machine learning algorithms. Among these combinations, MedImageInsight embeddings paired with an support vector machine adapter yielded the highest mean area under the curve (mAUC) at 93.8%, followed closely by Rad-DINO (91.1%) and CXR-Foundation (89.0%). In comparison, BiomedCLIP and DenseNet121 exhibited moderate performance with mAUC scores of 83.0% and 81.8%, respectively, whereas Med-Flamingo delivered the lowest performance at 75.1%. Notably, most adapter models demonstrated computational efficiency, achieving training within one minute and inference within seconds on CPU, underscoring their practicality for clinical applications. Furthermore, fairness analyses on adapters trained on MedImageInsight-derived embeddings indicated minimal disparities, with gender differences in performance within 2% and standard deviations across age groups not exceeding 3%. These findings confirm that foundation model embeddings-especially those from MedImageInsight-facilitate accurate, computationally efficient, and equitable diagnostic classification using lightweight adapters for radiographic image analysis.
MedImageInsight: An Open-Source Embedding Model for General Domain Medical Imaging
Oct 09, 2024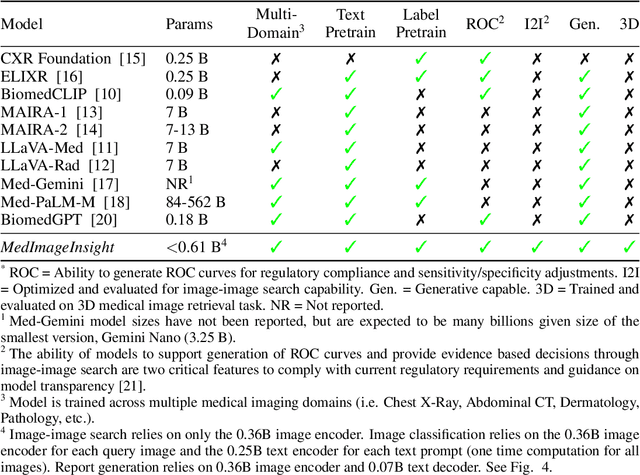
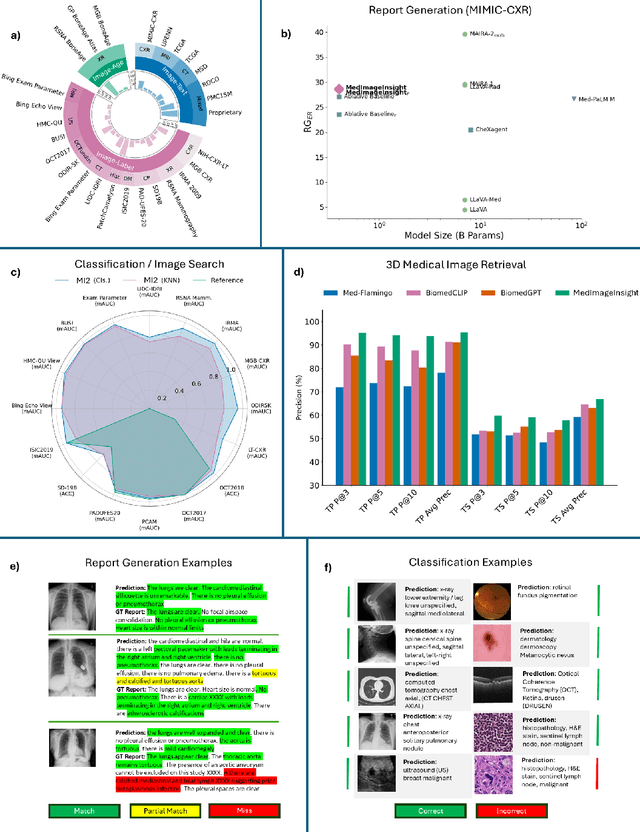
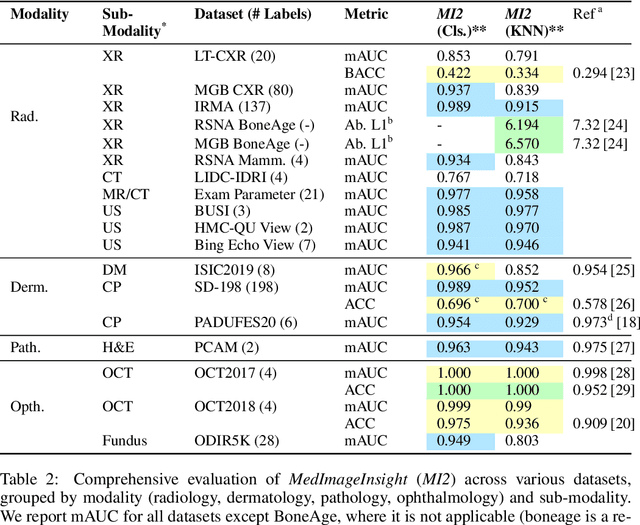
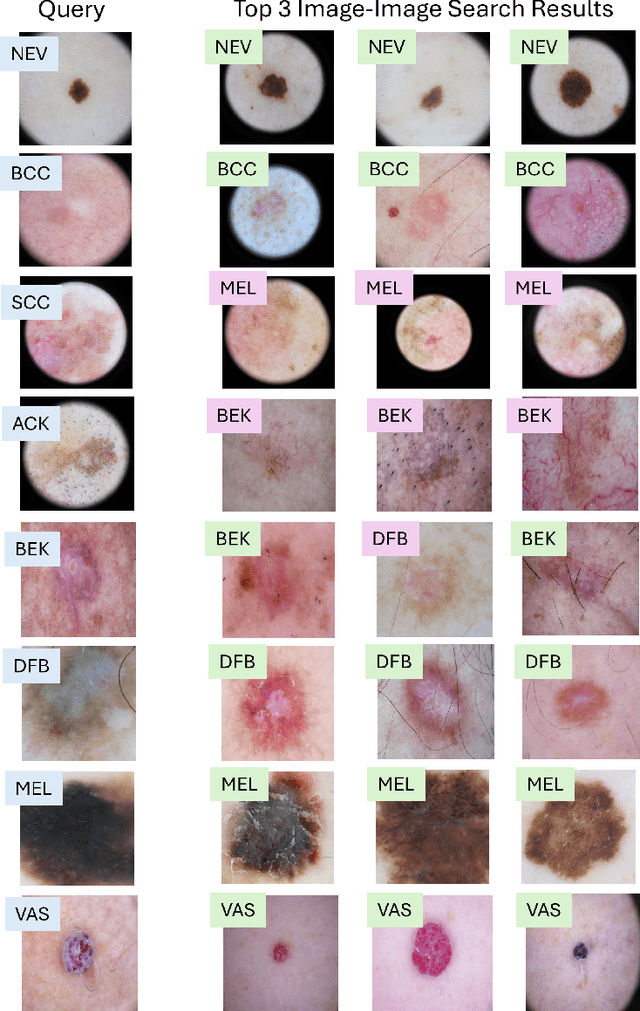
Abstract:In this work, we present MedImageInsight, an open-source medical imaging embedding model. MedImageInsight is trained on medical images with associated text and labels across a diverse collection of domains, including X-Ray, CT, MRI, dermoscopy, OCT, fundus photography, ultrasound, histopathology, and mammography. Rigorous evaluations demonstrate MedImageInsight's ability to achieve state-of-the-art (SOTA) or human expert level performance across classification, image-image search, and fine-tuning tasks. Specifically, on public datasets, MedImageInsight achieves SOTA in CT 3D medical image retrieval, as well as SOTA in disease classification and search for chest X-ray, dermatology, and OCT imaging. Furthermore, MedImageInsight achieves human expert performance in bone age estimation (on both public and partner data), as well as AUC above 0.9 in most other domains. When paired with a text decoder, MedImageInsight achieves near SOTA level single image report findings generation with less than 10\% the parameters of other models. Compared to fine-tuning GPT-4o with only MIMIC-CXR data for the same task, MedImageInsight outperforms in clinical metrics, but underperforms on lexical metrics where GPT-4o sets a new SOTA. Importantly for regulatory purposes, MedImageInsight can generate ROC curves, adjust sensitivity and specificity based on clinical need, and provide evidence-based decision support through image-image search (which can also enable retrieval augmented generation). In an independent clinical evaluation of image-image search in chest X-ray, MedImageInsight outperformed every other publicly available foundation model evaluated by large margins (over 6 points AUC), and significantly outperformed other models in terms of AI fairness (across age and gender). We hope releasing MedImageInsight will help enhance collective progress in medical imaging AI research and development.
A New Benchmark for Evaluation of Cross-Domain Few-Shot Learning
Dec 16, 2019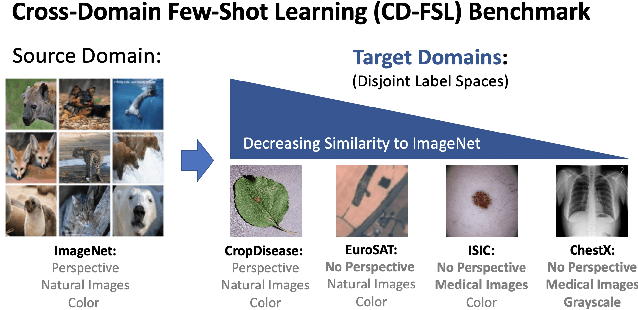
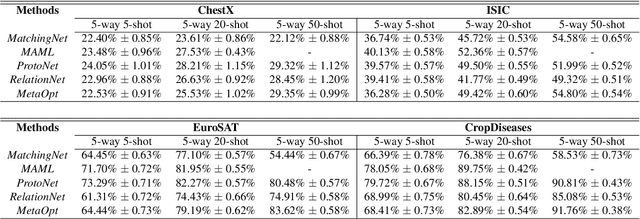
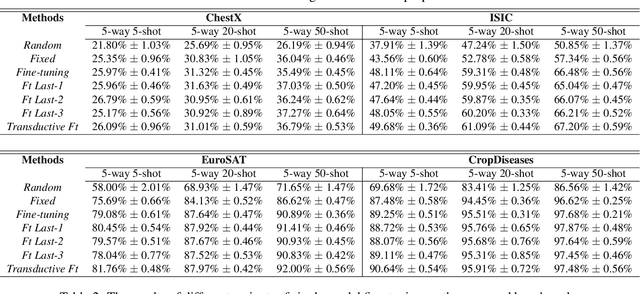
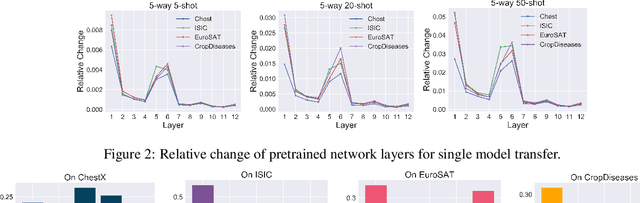
Abstract:Recent progress on few-shot learning has largely re-lied on annotated data for meta-learning, sampled from the same domain as the novel classes. However, in many applications, collecting data for meta-learning is infeasible or impossible. This leads to the cross-domain few-shot learn-ing problem, where a large domain shift exists between base and novel classes. Although some preliminary investigation of the few-shot methods under domain shift exists, a standard benchmark for cross-domain few-shot learning is not yet established. In this paper, we propose the cross-domain few-shot learning (CD-FSL) benchmark, consist-ing of images from diverse domains with varying similarity to ImageNet, ranging from crop disease images, satellite images, and medical images. Extensive experiments on the proposed benchmark are performed to compare an array of state-of-art meta-learning and transfer learning approaches, including various forms of single model fine-tuning and ensemble learning. The results demonstrate that current meta-learning methods underperform in relation to simple fine-tuning by 12.8% average accuracy. Accuracy of all methods tend to correlate with dataset similarity toImageNet. In addition, the relative performance gain with increasing number of shots is greater with transfer methods compared to meta-learning. Finally, we demonstrate that transferring from multiple pretrained models achieves best performance, with accuracy improvements of 14.9% and 1.9% versus the best of meta-learning and single model fine-tuning approaches, respectively. In summary, the proposed benchmark serves as a challenging platform to guide future research on cross-domain few-shot learning due to its spectrum of diversity and coverage
Estimating Skin Tone and Effects on Classification Performance in Dermatology Datasets
Oct 29, 2019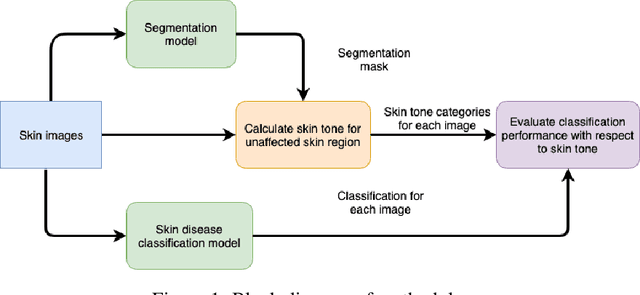
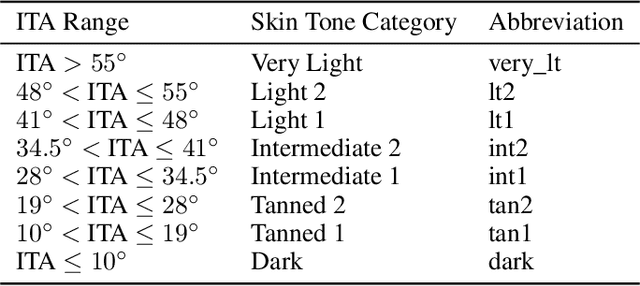
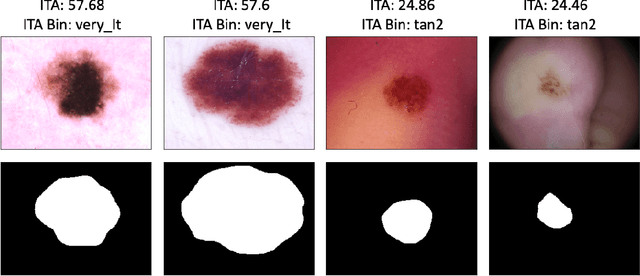

Abstract:Recent advances in computer vision and deep learning have led to breakthroughs in the development of automated skin image analysis. In particular, skin cancer classification models have achieved performance higher than trained expert dermatologists. However, no attempt has been made to evaluate the consistency in performance of machine learning models across populations with varying skin tones. In this paper, we present an approach to estimate skin tone in benchmark skin disease datasets, and investigate whether model performance is dependent on this measure. Specifically, we use individual typology angle (ITA) to approximate skin tone in dermatology datasets. We look at the distribution of ITA values to better understand skin color representation in two benchmark datasets: 1) the ISIC 2018 Challenge dataset, a collection of dermoscopic images of skin lesions for the detection of skin cancer, and 2) the SD-198 dataset, a collection of clinical images capturing a wide variety of skin diseases. To estimate ITA, we first develop segmentation models to isolate non-diseased areas of skin. We find that the majority of the data in the the two datasets have ITA values between 34.5{\deg} and 48{\deg}, which are associated with lighter skin, and is consistent with under-representation of darker skinned populations in these datasets. We also find no measurable correlation between performance of machine learning model and ITA values, though more comprehensive data is needed for further validation.
BCN20000: Dermoscopic Lesions in the Wild
Aug 30, 2019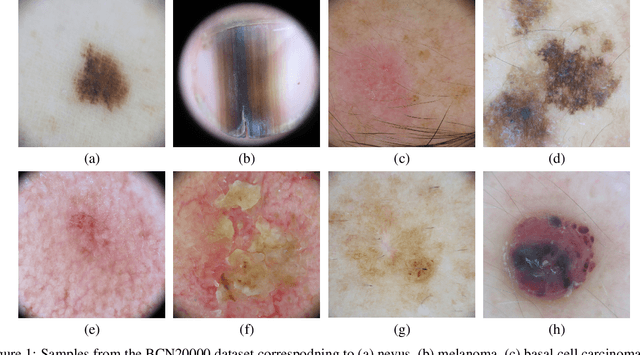
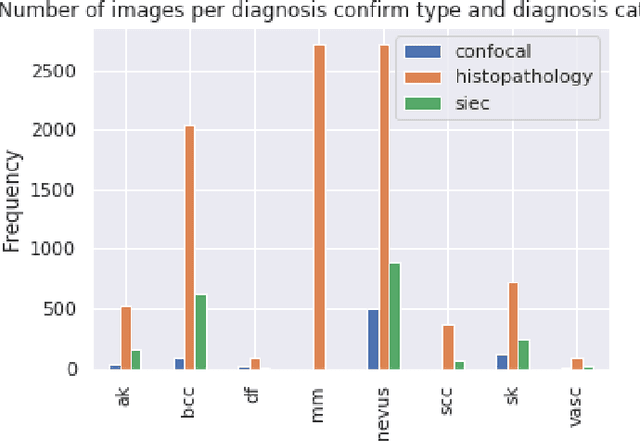
Abstract:This article summarizes the BCN20000 dataset, composed of 19424 dermoscopic images of skin lesions captured from 2010 to 2016 in the facilities of the Hospital Cl\'inic in Barcelona. With this dataset, we aim to study the problem of unconstrained classification of dermoscopic images of skin cancer, including lesions found in hard-to-diagnose locations (nails and mucosa), large lesions which do not fit in the aperture of the dermoscopy device, and hypo-pigmented lesions. The BCN20000 will be provided to the participants of the ISIC Challenge 2019, where they will be asked to train algorithms to classify dermoscopic images of skin cancer automatically.
Teaching AI to Explain its Decisions Using Embeddings and Multi-Task Learning
Jun 05, 2019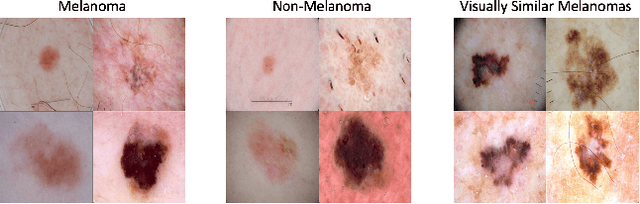
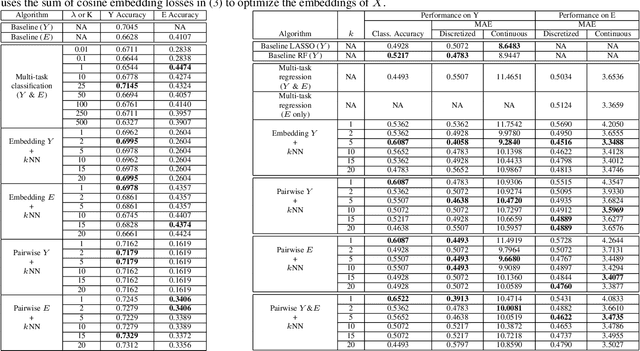
Abstract:Using machine learning in high-stakes applications often requires predictions to be accompanied by explanations comprehensible to the domain user, who has ultimate responsibility for decisions and outcomes. Recently, a new framework for providing explanations, called TED, has been proposed to provide meaningful explanations for predictions. This framework augments training data to include explanations elicited from domain users, in addition to features and labels. This approach ensures that explanations for predictions are tailored to the complexity expectations and domain knowledge of the consumer. In this paper, we build on this foundational work, by exploring more sophisticated instantiations of the TED framework and empirically evaluate their effectiveness in two diverse domains, chemical odor and skin cancer prediction. Results demonstrate that meaningful explanations can be reliably taught to machine learning algorithms, and in some cases, improving modeling accuracy.
TED: Teaching AI to Explain its Decisions
Nov 12, 2018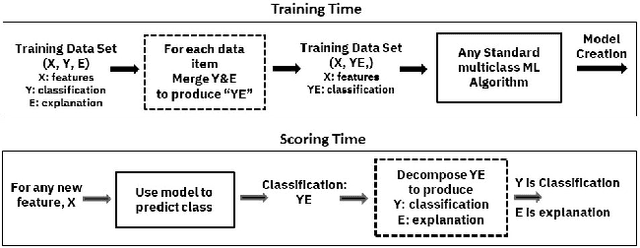
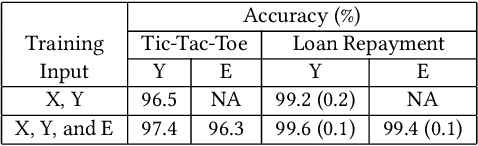
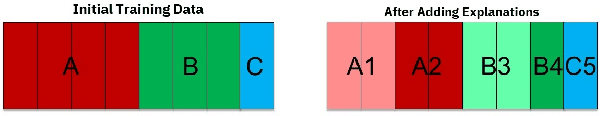
Abstract:Artificial intelligence systems are being increasingly deployed due to their potential to increase the efficiency, scale, consistency, fairness, and accuracy of decisions. However, as many of these systems are opaque in their operation, there is a growing demand for such systems to provide explanations for their decisions. Conventional approaches to this problem attempt to expose or discover the inner workings of a machine learning model with the hope that the resulting explanations will be meaningful to the consumer. In contrast, this paper suggests a new approach to this problem. It introduces a simple, practical framework, called Teaching Explanations for Decisions (TED), that provides meaningful explanations that match the mental model of the consumer. We illustrate the generality and effectiveness of this approach with two different examples, resulting in highly accurate explanations with no loss of prediction accuracy for these two examples.
Teaching Meaningful Explanations
Sep 11, 2018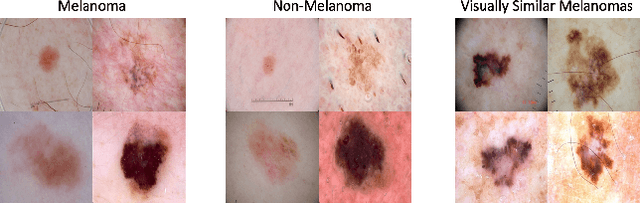
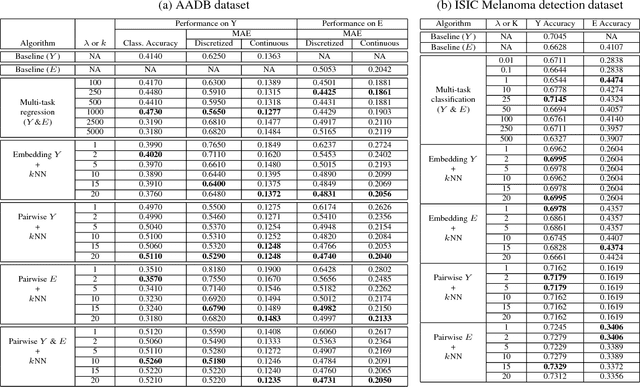
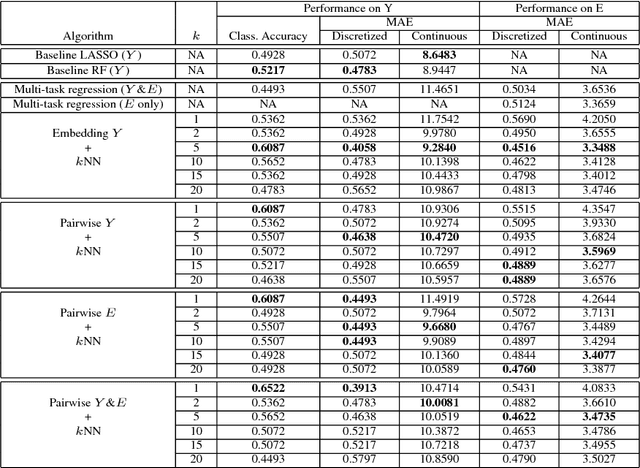
Abstract:The adoption of machine learning in high-stakes applications such as healthcare and law has lagged in part because predictions are not accompanied by explanations comprehensible to the domain user, who often holds the ultimate responsibility for decisions and outcomes. In this paper, we propose an approach to generate such explanations in which training data is augmented to include, in addition to features and labels, explanations elicited from domain users. A joint model is then learned to produce both labels and explanations from the input features. This simple idea ensures that explanations are tailored to the complexity expectations and domain knowledge of the consumer. Evaluation spans multiple modeling techniques on a game dataset, a (visual) aesthetics dataset, a chemical odor dataset and a Melanoma dataset showing that our approach is generalizable across domains and algorithms. Results demonstrate that meaningful explanations can be reliably taught to machine learning algorithms, and in some cases, also improve modeling accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge