Veronica Rotemberg
On the Role of Calibration in Benchmarking Algorithmic Fairness for Skin Cancer Detection
Nov 10, 2025Abstract:Artificial Intelligence (AI) models have demonstrated expert-level performance in melanoma detection, yet their clinical adoption is hindered by performance disparities across demographic subgroups such as gender, race, and age. Previous efforts to benchmark the performance of AI models have primarily focused on assessing model performance using group fairness metrics that rely on the Area Under the Receiver Operating Characteristic curve (AUROC), which does not provide insights into a model's ability to provide accurate estimates. In line with clinical assessments, this paper addresses this gap by incorporating calibration as a complementary benchmarking metric to AUROC-based fairness metrics. Calibration evaluates the alignment between predicted probabilities and observed event rates, offering deeper insights into subgroup biases. We assess the performance of the leading skin cancer detection algorithm of the ISIC 2020 Challenge on the ISIC 2020 Challenge dataset and the PROVE-AI dataset, and compare it with the second and third place models, focusing on subgroups defined by sex, race (Fitzpatrick Skin Tone), and age. Our findings reveal that while existing models enhance discriminative accuracy, they often over-diagnose risk and exhibit calibration issues when applied to new datasets. This study underscores the necessity for comprehensive model auditing strategies and extensive metadata collection to achieve equitable AI-driven healthcare solutions. All code is publicly available at https://github.com/bdominique/testing_strong_calibration.
* 19 pages, 4 figures. Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:027
Augmenting medical image classifiers with synthetic data from latent diffusion models
Aug 23, 2023



Abstract:While hundreds of artificial intelligence (AI) algorithms are now approved or cleared by the US Food and Drugs Administration (FDA), many studies have shown inconsistent generalization or latent bias, particularly for underrepresented populations. Some have proposed that generative AI could reduce the need for real data, but its utility in model development remains unclear. Skin disease serves as a useful case study in synthetic image generation due to the diversity of disease appearance, particularly across the protected attribute of skin tone. Here we show that latent diffusion models can scalably generate images of skin disease and that augmenting model training with these data improves performance in data-limited settings. These performance gains saturate at synthetic-to-real image ratios above 10:1 and are substantially smaller than the gains obtained from adding real images. As part of our analysis, we generate and analyze a new dataset of 458,920 synthetic images produced using several generation strategies. Our results suggest that synthetic data could serve as a force-multiplier for model development, but the collection of diverse real-world data remains the most important step to improve medical AI algorithms.
Disparities in Dermatology AI Performance on a Diverse, Curated Clinical Image Set
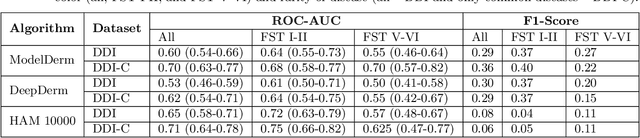
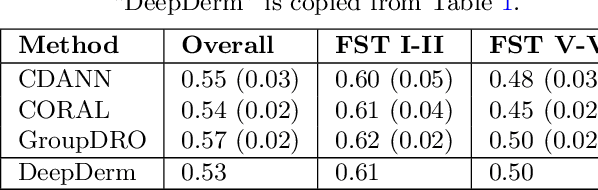
Mar 15, 2022Abstract:Access to dermatological care is a major issue, with an estimated 3 billion people lacking access to care globally. Artificial intelligence (AI) may aid in triaging skin diseases. However, most AI models have not been rigorously assessed on images of diverse skin tones or uncommon diseases. To ascertain potential biases in algorithm performance in this context, we curated the Diverse Dermatology Images (DDI) dataset-the first publicly available, expertly curated, and pathologically confirmed image dataset with diverse skin tones. Using this dataset of 656 images, we show that state-of-the-art dermatology AI models perform substantially worse on DDI, with receiver operator curve area under the curve (ROC-AUC) dropping by 27-36 percent compared to the models' original test results. All the models performed worse on dark skin tones and uncommon diseases, which are represented in the DDI dataset. Additionally, we find that dermatologists, who typically provide visual labels for AI training and test datasets, also perform worse on images of dark skin tones and uncommon diseases compared to ground truth biopsy annotations. Finally, fine-tuning AI models on the well-characterized and diverse DDI images closed the performance gap between light and dark skin tones. Moreover, algorithms fine-tuned on diverse skin tones outperformed dermatologists on identifying malignancy on images of dark skin tones. Our findings identify important weaknesses and biases in dermatology AI that need to be addressed to ensure reliable application to diverse patients and diseases.
Disparities in Dermatology AI: Assessments Using Diverse Clinical Images
Nov 15, 2021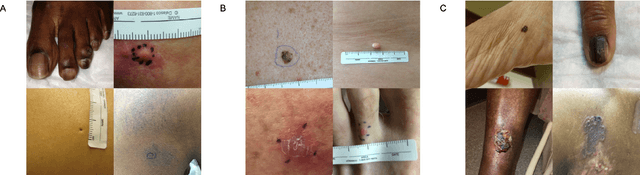
Abstract:More than 3 billion people lack access to care for skin disease. AI diagnostic tools may aid in early skin cancer detection; however most models have not been assessed on images of diverse skin tones or uncommon diseases. To address this, we curated the Diverse Dermatology Images (DDI) dataset - the first publicly available, pathologically confirmed images featuring diverse skin tones. We show that state-of-the-art dermatology AI models perform substantially worse on DDI, with ROC-AUC dropping 29-40 percent compared to the models' original results. We find that dark skin tones and uncommon diseases, which are well represented in the DDI dataset, lead to performance drop-offs. Additionally, we show that state-of-the-art robust training methods cannot correct for these biases without diverse training data. Our findings identify important weaknesses and biases in dermatology AI that need to be addressed to ensure reliable application to diverse patients and across all disease.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020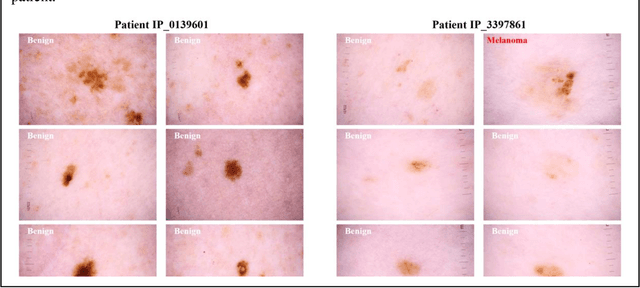
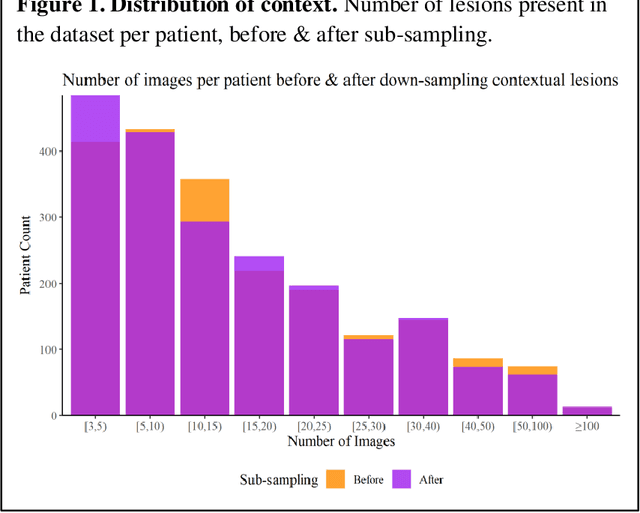
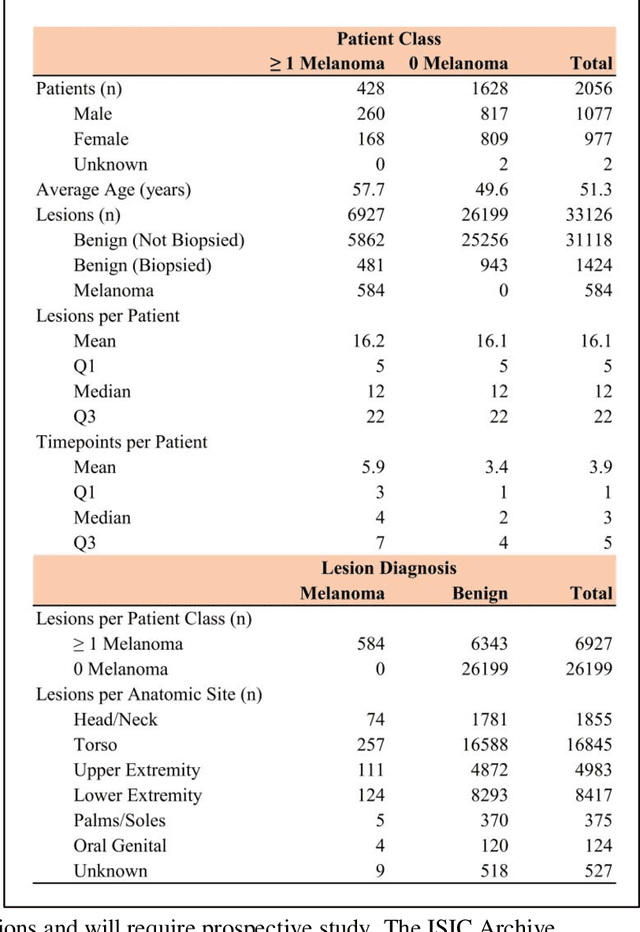
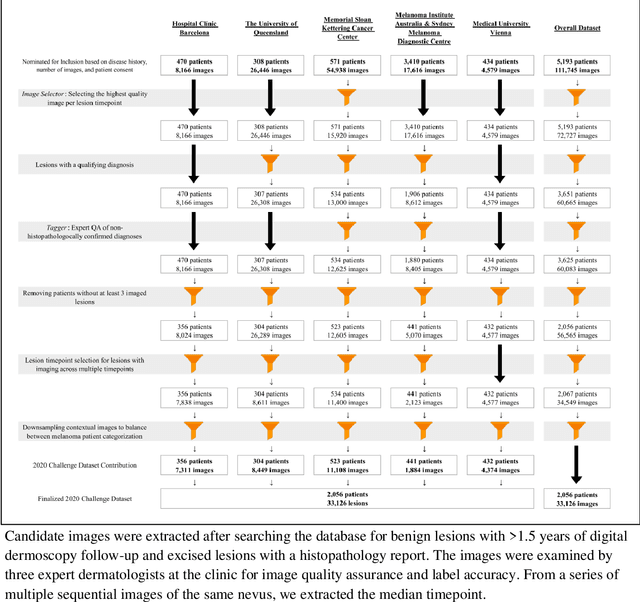
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
BCN20000: Dermoscopic Lesions in the Wild
Aug 30, 2019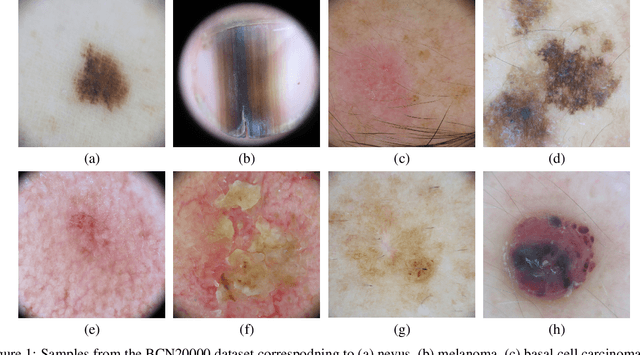
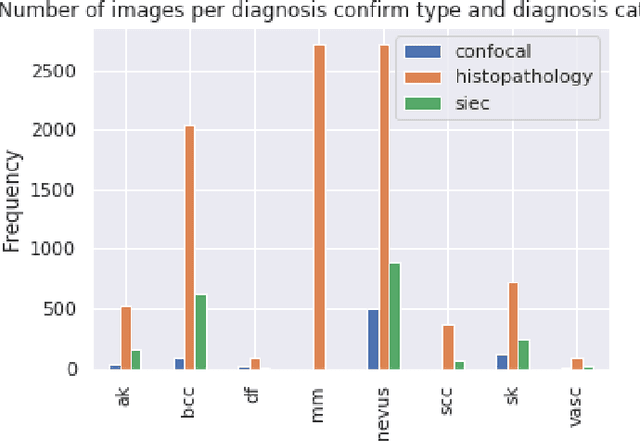
Abstract:This article summarizes the BCN20000 dataset, composed of 19424 dermoscopic images of skin lesions captured from 2010 to 2016 in the facilities of the Hospital Cl\'inic in Barcelona. With this dataset, we aim to study the problem of unconstrained classification of dermoscopic images of skin cancer, including lesions found in hard-to-diagnose locations (nails and mucosa), large lesions which do not fit in the aperture of the dermoscopy device, and hypo-pigmented lesions. The BCN20000 will be provided to the participants of the ISIC Challenge 2019, where they will be asked to train algorithms to classify dermoscopic images of skin cancer automatically.
Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)
Mar 29, 2019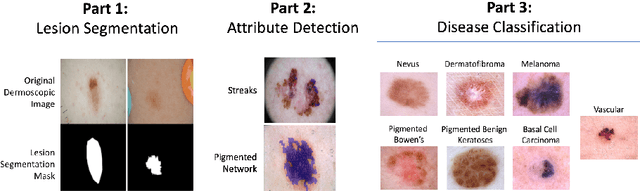
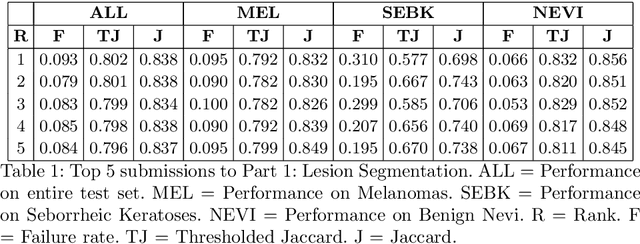
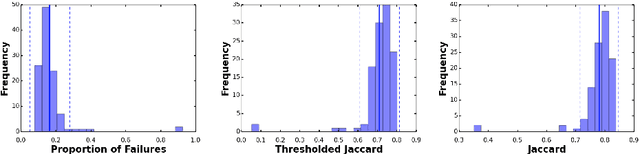
Abstract:This work summarizes the results of the largest skin image analysis challenge in the world, hosted by the International Skin Imaging Collaboration (ISIC), a global partnership that has organized the world's largest public repository of dermoscopic images of skin. The challenge was hosted in 2018 at the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference in Granada, Spain. The dataset included over 12,500 images across 3 tasks. 900 users registered for data download, 115 submitted to the lesion segmentation task, 25 submitted to the lesion attribute detection task, and 159 submitted to the disease classification task. Novel evaluation protocols were established, including a new test for segmentation algorithm performance, and a test for algorithm ability to generalize. Results show that top segmentation algorithms still fail on over 10% of images on average, and algorithms with equal performance on test data can have different abilities to generalize. This is an important consideration for agencies regulating the growing set of machine learning tools in the healthcare domain, and sets a new standard for future public challenges in healthcare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge