Brigid Betz-Stablein
Prompt-driven Latent Domain Generalization for Medical Image Classification
Jan 05, 2024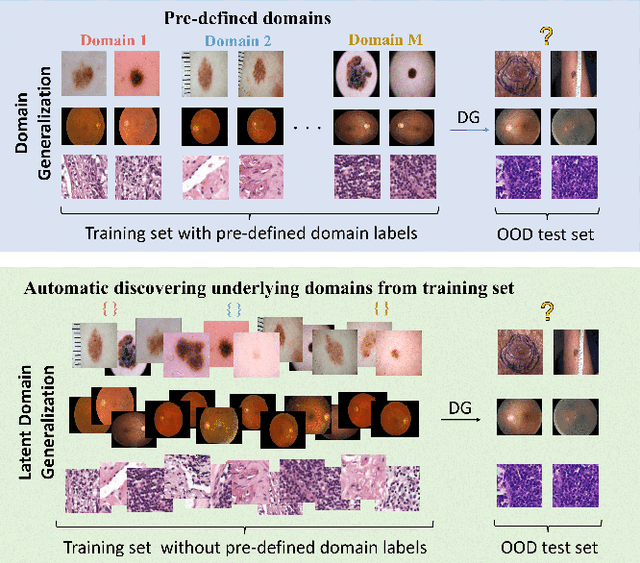
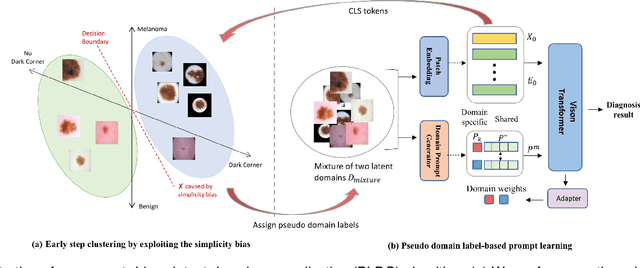
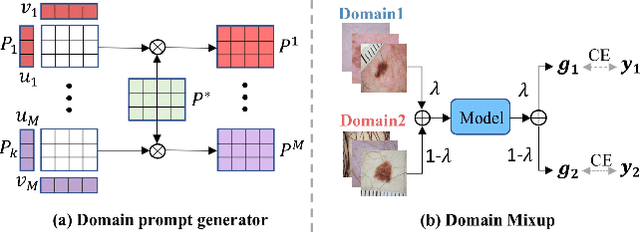
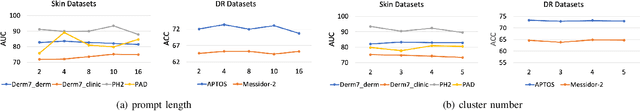
Abstract:Deep learning models for medical image analysis easily suffer from distribution shifts caused by dataset artifacts bias, camera variations, differences in the imaging station, etc., leading to unreliable diagnoses in real-world clinical settings. Domain generalization (DG) methods, which aim to train models on multiple domains to perform well on unseen domains, offer a promising direction to solve the problem. However, existing DG methods assume domain labels of each image are available and accurate, which is typically feasible for only a limited number of medical datasets. To address these challenges, we propose a novel DG framework for medical image classification without relying on domain labels, called Prompt-driven Latent Domain Generalization (PLDG). PLDG consists of unsupervised domain discovery and prompt learning. This framework first discovers pseudo domain labels by clustering the bias-associated style features, then leverages collaborative domain prompts to guide a Vision Transformer to learn knowledge from discovered diverse domains. To facilitate cross-domain knowledge learning between different prompts, we introduce a domain prompt generator that enables knowledge sharing between domain prompts and a shared prompt. A domain mixup strategy is additionally employed for more flexible decision margins and mitigates the risk of incorrect domain assignments. Extensive experiments on three medical image classification tasks and one debiasing task demonstrate that our method can achieve comparable or even superior performance than conventional DG algorithms without relying on domain labels. Our code will be publicly available upon the paper is accepted.
Revamping AI Models in Dermatology: Overcoming Critical Challenges for Enhanced Skin Lesion Diagnosis
Nov 02, 2023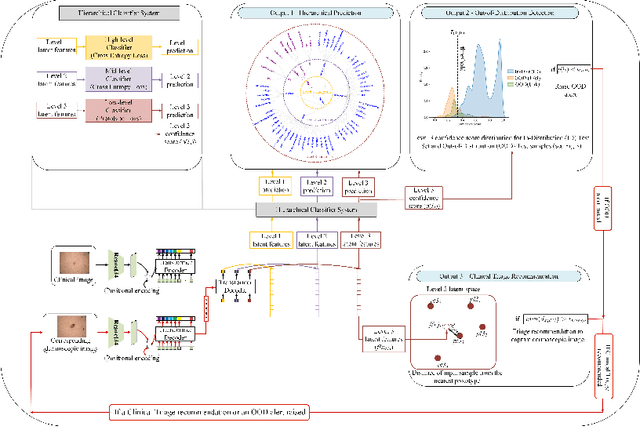
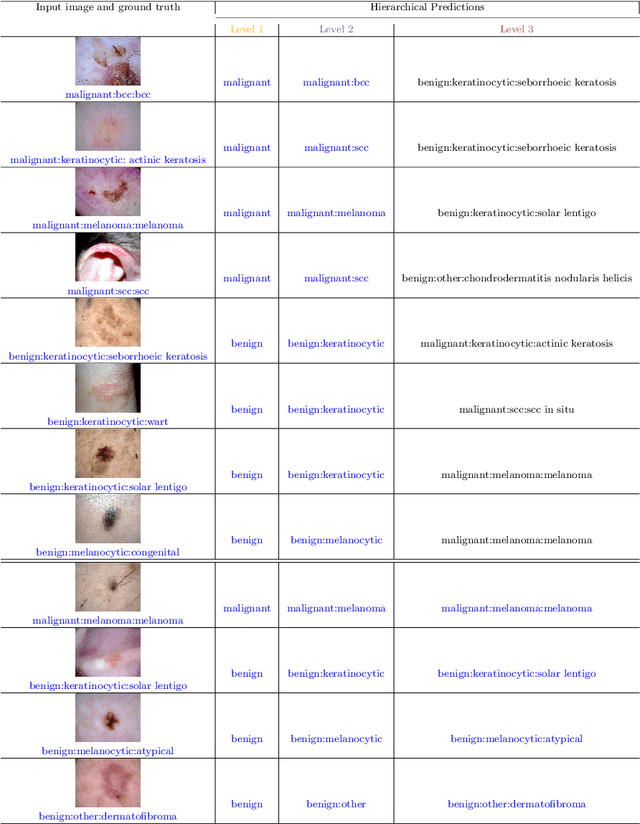
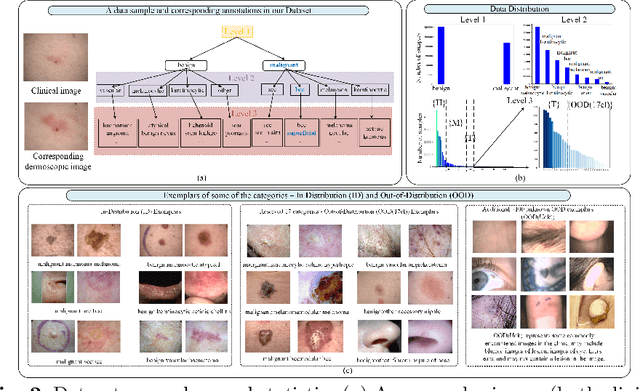

Abstract:The surge in developing deep learning models for diagnosing skin lesions through image analysis is notable, yet their clinical black faces challenges. Current dermatology AI models have limitations: limited number of possible diagnostic outputs, lack of real-world testing on uncommon skin lesions, inability to detect out-of-distribution images, and over-reliance on dermoscopic images. To address these, we present an All-In-One \textbf{H}ierarchical-\textbf{O}ut of Distribution-\textbf{C}linical Triage (HOT) model. For a clinical image, our model generates three outputs: a hierarchical prediction, an alert for out-of-distribution images, and a recommendation for dermoscopy if clinical image alone is insufficient for diagnosis. When the recommendation is pursued, it integrates both clinical and dermoscopic images to deliver final diagnosis. Extensive experiments on a representative cutaneous lesion dataset demonstrate the effectiveness and synergy of each component within our framework. Our versatile model provides valuable decision support for lesion diagnosis and sets a promising precedent for medical AI applications.
Ugly Ducklings or Swans: A Tiered Quadruplet Network with Patient-Specific Mining for Improved Skin Lesion Classification
Sep 18, 2023Abstract:An ugly duckling is an obviously different skin lesion from surrounding lesions of an individual, and the ugly duckling sign is a criterion used to aid in the diagnosis of cutaneous melanoma by differentiating between highly suspicious and benign lesions. However, the appearance of pigmented lesions, can change drastically from one patient to another, resulting in difficulties in visual separation of ugly ducklings. Hence, we propose DMT-Quadruplet - a deep metric learning network to learn lesion features at two tiers - patient-level and lesion-level. We introduce a patient-specific quadruplet mining approach together with a tiered quadruplet network, to drive the network to learn more contextual information both globally and locally between the two tiers. We further incorporate a dynamic margin within the patient-specific mining to allow more useful quadruplets to be mined within individuals. Comprehensive experiments show that our proposed method outperforms traditional classifiers, achieving 54% higher sensitivity than a baseline ResNet18 CNN and 37% higher than a naive triplet network in classifying ugly duckling lesions. Visualisation of the data manifold in the metric space further illustrates that DMT-Quadruplet is capable of classifying ugly duckling lesions in both patient-specific and patient-agnostic manner successfully.
Application of Machine Learning in Melanoma Detection and the Identification of 'Ugly Duckling' and Suspicious Naevi: A Review
Sep 05, 2023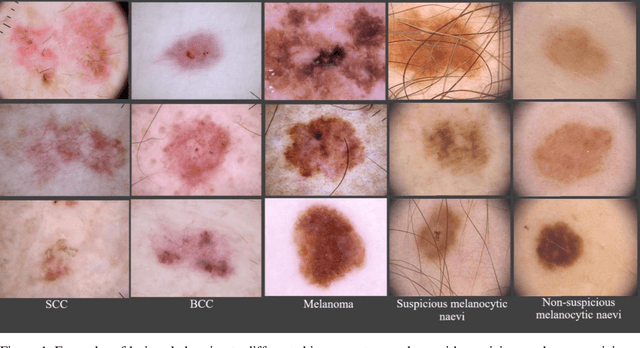
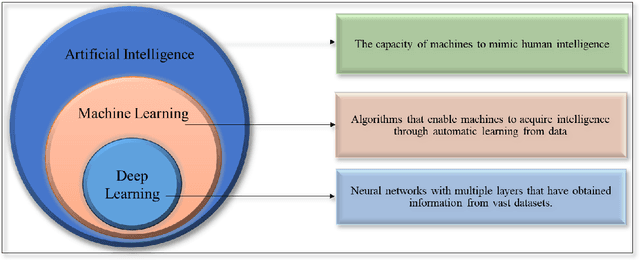

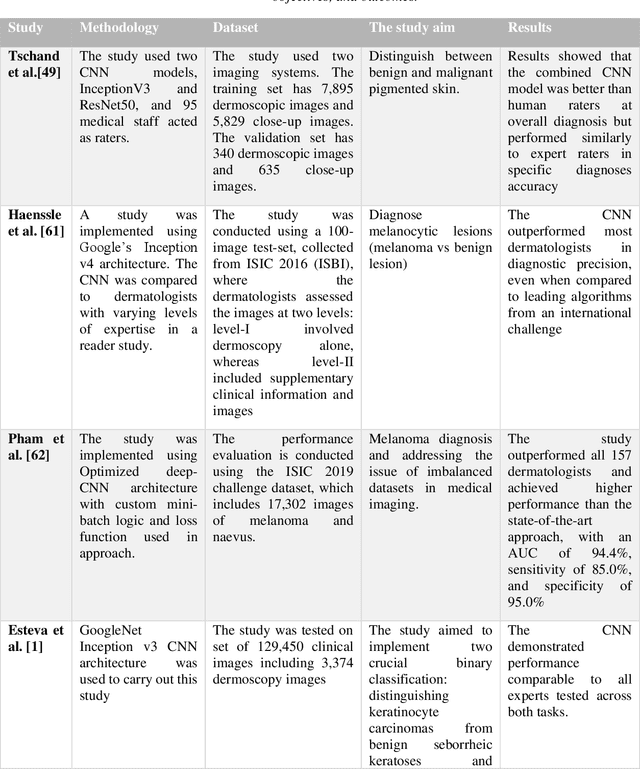
Abstract:Skin lesions known as naevi exhibit diverse characteristics such as size, shape, and colouration. The concept of an "Ugly Duckling Naevus" comes into play when monitoring for melanoma, referring to a lesion with distinctive features that sets it apart from other lesions in the vicinity. As lesions within the same individual typically share similarities and follow a predictable pattern, an ugly duckling naevus stands out as unusual and may indicate the presence of a cancerous melanoma. Computer-aided diagnosis (CAD) has become a significant player in the research and development field, as it combines machine learning techniques with a variety of patient analysis methods. Its aim is to increase accuracy and simplify decision-making, all while responding to the shortage of specialized professionals. These automated systems are especially important in skin cancer diagnosis where specialist availability is limited. As a result, their use could lead to life-saving benefits and cost reductions within healthcare. Given the drastic change in survival when comparing early stage to late-stage melanoma, early detection is vital for effective treatment and patient outcomes. Machine learning (ML) and deep learning (DL) techniques have gained popularity in skin cancer classification, effectively addressing challenges, and providing results equivalent to that of specialists. This article extensively covers modern Machine Learning and Deep Learning algorithms for detecting melanoma and suspicious naevi. It begins with general information on skin cancer and different types of naevi, then introduces AI, ML, DL, and CAD. The article then discusses the successful applications of various ML techniques like convolutional neural networks (CNN) for melanoma detection compared to dermatologists' performance. Lastly, it examines ML methods for UD naevus detection and identifying suspicious naevi.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020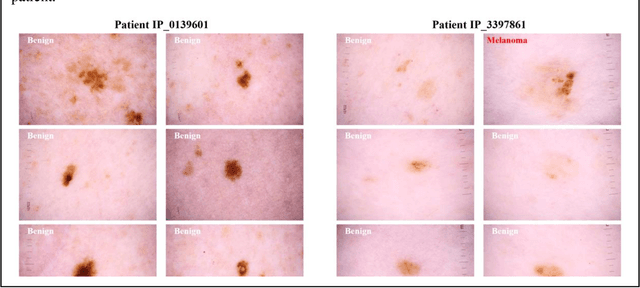
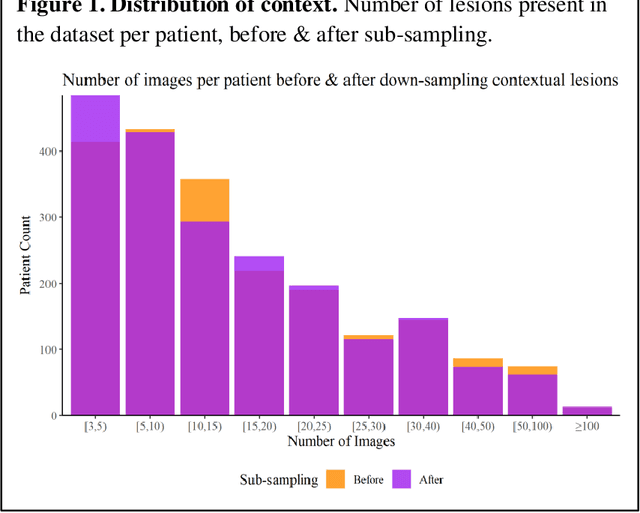
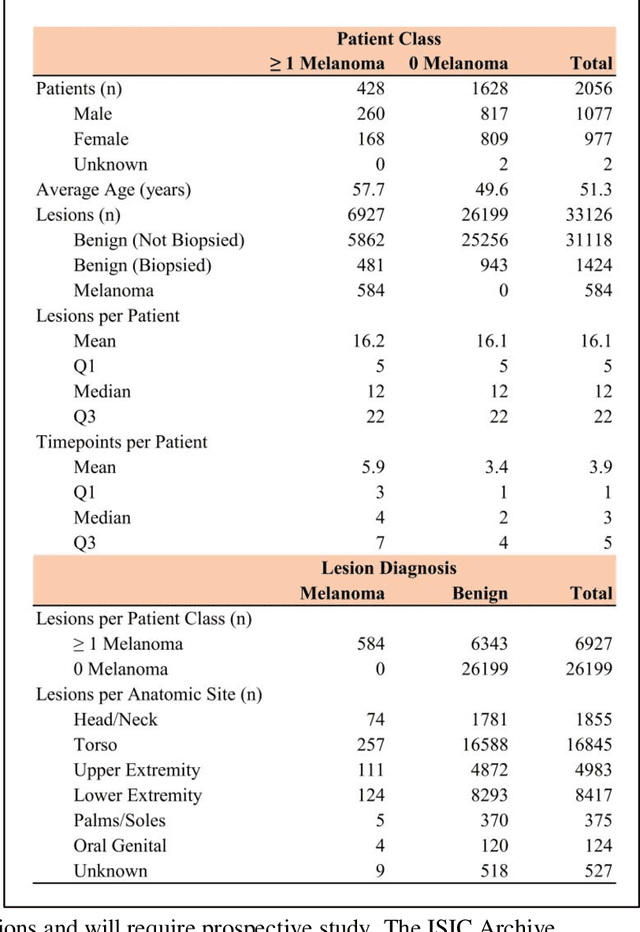
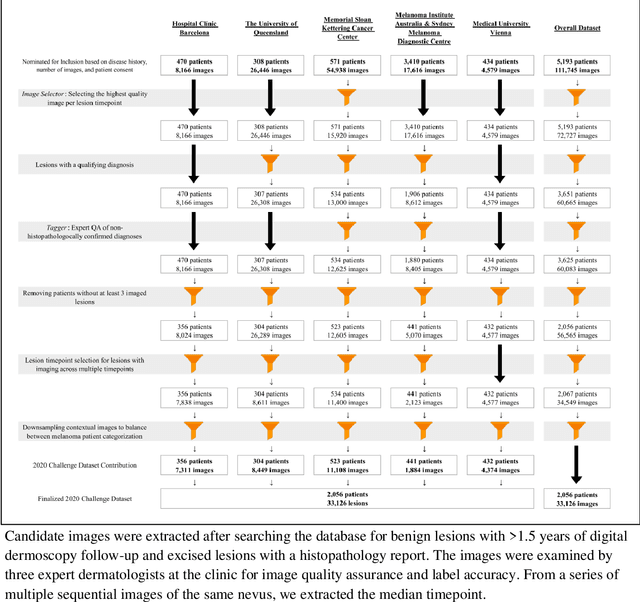
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge