Zhen Yu
Renmin University of China
Controllable Skin Synthesis via Lesion-Focused Vector Autoregression Model
Aug 27, 2025Abstract:Skin images from real-world clinical practice are often limited, resulting in a shortage of training data for deep-learning models. While many studies have explored skin image synthesis, existing methods often generate low-quality images and lack control over the lesion's location and type. To address these limitations, we present LF-VAR, a model leveraging quantified lesion measurement scores and lesion type labels to guide the clinically relevant and controllable synthesis of skin images. It enables controlled skin synthesis with specific lesion characteristics based on language prompts. We train a multiscale lesion-focused Vector Quantised Variational Auto-Encoder (VQVAE) to encode images into discrete latent representations for structured tokenization. Then, a Visual AutoRegressive (VAR) Transformer trained on tokenized representations facilitates image synthesis. Lesion measurement from the lesion region and types as conditional embeddings are integrated to enhance synthesis fidelity. Our method achieves the best overall FID score (average 0.74) among seven lesion types, improving upon the previous state-of-the-art (SOTA) by 6.3%. The study highlights our controllable skin synthesis model's effectiveness in generating high-fidelity, clinically relevant synthetic skin images. Our framework code is available at https://github.com/echosun1996/LF-VAR.
MAKE: Multi-Aspect Knowledge-Enhanced Vision-Language Pretraining for Zero-shot Dermatological Assessment
May 14, 2025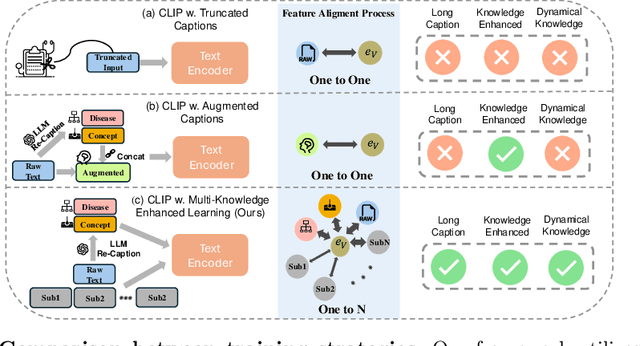
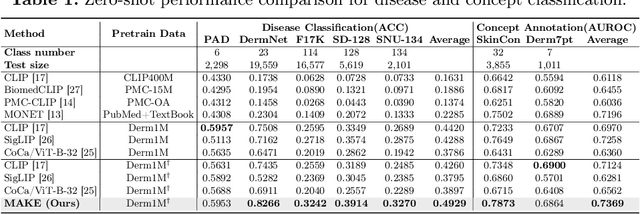
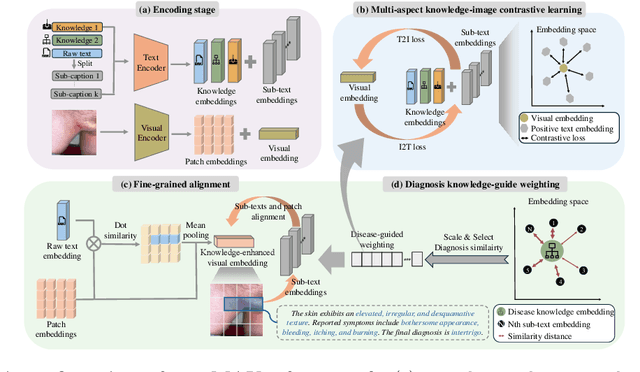
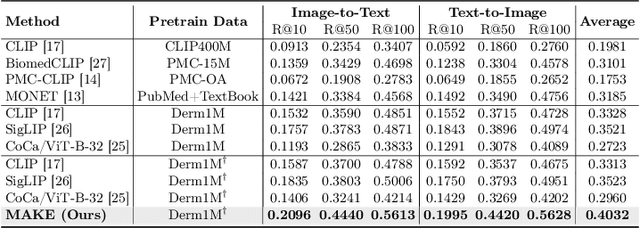
Abstract:Dermatological diagnosis represents a complex multimodal challenge that requires integrating visual features with specialized clinical knowledge. While vision-language pretraining (VLP) has advanced medical AI, its effectiveness in dermatology is limited by text length constraints and the lack of structured texts. In this paper, we introduce MAKE, a Multi-Aspect Knowledge-Enhanced vision-language pretraining framework for zero-shot dermatological tasks. Recognizing that comprehensive dermatological descriptions require multiple knowledge aspects that exceed standard text constraints, our framework introduces: (1) a multi-aspect contrastive learning strategy that decomposes clinical narratives into knowledge-enhanced sub-texts through large language models, (2) a fine-grained alignment mechanism that connects subcaptions with diagnostically relevant image features, and (3) a diagnosis-guided weighting scheme that adaptively prioritizes different sub-captions based on clinical significance prior. Through pretraining on 403,563 dermatological image-text pairs collected from education resources, MAKE significantly outperforms state-of-the-art VLP models on eight datasets across zero-shot skin disease classification, concept annotation, and cross-modal retrieval tasks. Our code will be made publicly available at https: //github.com/SiyuanYan1/MAKE.
scDD: Latent Codes Based scRNA-seq Dataset Distillation with Foundation Model Knowledge
Mar 06, 2025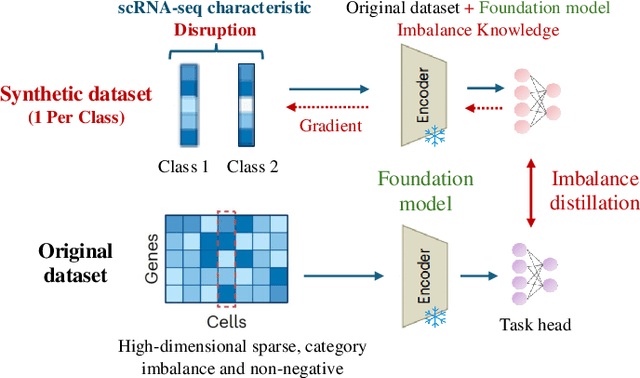
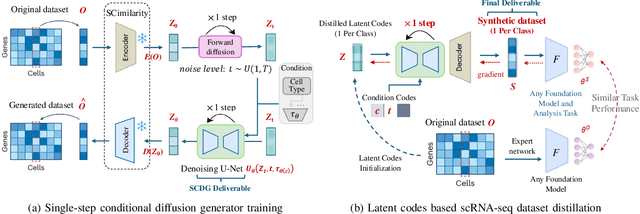
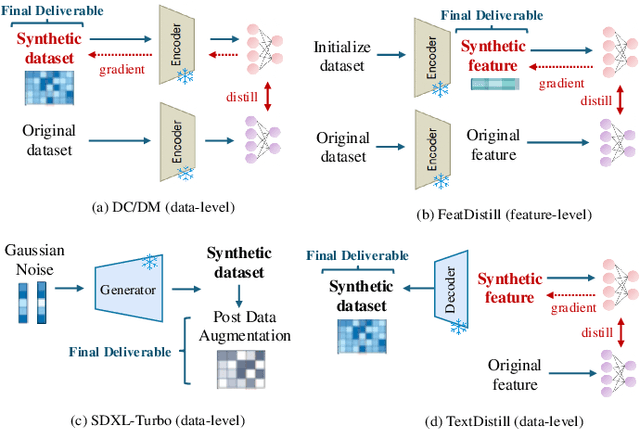
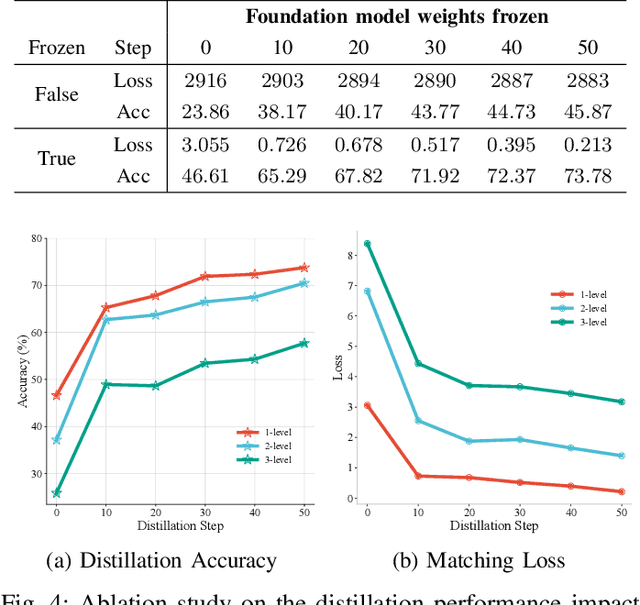
Abstract:Single-cell RNA sequencing (scRNA-seq) technology has profiled hundreds of millions of human cells across organs, diseases, development and perturbations to date. However, the high-dimensional sparsity, batch effect noise, category imbalance, and ever-increasing data scale of the original sequencing data pose significant challenges for multi-center knowledge transfer, data fusion, and cross-validation between scRNA-seq datasets. To address these barriers, (1) we first propose a latent codes-based scRNA-seq dataset distillation framework named scDD, which transfers and distills foundation model knowledge and original dataset information into a compact latent space and generates synthetic scRNA-seq dataset by a generator to replace the original dataset. Then, (2) we propose a single-step conditional diffusion generator named SCDG, which perform single-step gradient back-propagation to help scDD optimize distillation quality and avoid gradient decay caused by multi-step back-propagation. Meanwhile, SCDG ensures the scRNA-seq data characteristics and inter-class discriminability of the synthetic dataset through flexible conditional control and generation quality assurance. Finally, we propose a comprehensive benchmark to evaluate the performance of scRNA-seq dataset distillation in different data analysis tasks. It is validated that our proposed method can achieve 7.61% absolute and 15.70% relative improvement over previous state-of-the-art methods on average task.
CtrlNeRF: The Generative Neural Radiation Fields for the Controllable Synthesis of High-fidelity 3D-Aware Images
Dec 01, 2024Abstract:The neural radiance field (NERF) advocates learning the continuous representation of 3D geometry through a multilayer perceptron (MLP). By integrating this into a generative model, the generative neural radiance field (GRAF) is capable of producing images from random noise z without 3D supervision. In practice, the shape and appearance are modeled by z_s and z_a, respectively, to manipulate them separately during inference. However, it is challenging to represent multiple scenes using a solitary MLP and precisely control the generation of 3D geometry in terms of shape and appearance. In this paper, we introduce a controllable generative model (i.e. \textbf{CtrlNeRF}) that uses a single MLP network to represent multiple scenes with shared weights. Consequently, we manipulated the shape and appearance codes to realize the controllable generation of high-fidelity images with 3D consistency. Moreover, the model enables the synthesis of novel views that do not exist in the training sets via camera pose alteration and feature interpolation. Extensive experiments were conducted to demonstrate its superiority in 3D-aware image generation compared to its counterparts.
A General-Purpose Multimodal Foundation Model for Dermatology
Oct 19, 2024



Abstract:Diagnosing and treating skin diseases require advanced visual skills across multiple domains and the ability to synthesize information from various imaging modalities. Current deep learning models, while effective at specific tasks such as diagnosing skin cancer from dermoscopic images, fall short in addressing the complex, multimodal demands of clinical practice. Here, we introduce PanDerm, a multimodal dermatology foundation model pretrained through self-supervised learning on a dataset of over 2 million real-world images of skin diseases, sourced from 11 clinical institutions across 4 imaging modalities. We evaluated PanDerm on 28 diverse datasets covering a range of clinical tasks, including skin cancer screening, phenotype assessment and risk stratification, diagnosis of neoplastic and inflammatory skin diseases, skin lesion segmentation, change monitoring, and metastasis prediction and prognosis. PanDerm achieved state-of-the-art performance across all evaluated tasks, often outperforming existing models even when using only 5-10% of labeled data. PanDerm's clinical utility was demonstrated through reader studies in real-world clinical settings across multiple imaging modalities. It outperformed clinicians by 10.2% in early-stage melanoma detection accuracy and enhanced clinicians' multiclass skin cancer diagnostic accuracy by 11% in a collaborative human-AI setting. Additionally, PanDerm demonstrated robust performance across diverse demographic factors, including different body locations, age groups, genders, and skin tones. The strong results in benchmark evaluations and real-world clinical scenarios suggest that PanDerm could enhance the management of skin diseases and serve as a model for developing multimodal foundation models in other medical specialties, potentially accelerating the integration of AI support in healthcare.
Adaptive Transformer Modelling of Density Function for Nonparametric Survival Analysis
Sep 10, 2024Abstract:Survival analysis holds a crucial role across diverse disciplines, such as economics, engineering and healthcare. It empowers researchers to analyze both time-invariant and time-varying data, encompassing phenomena like customer churn, material degradation and various medical outcomes. Given the complexity and heterogeneity of such data, recent endeavors have demonstrated successful integration of deep learning methodologies to address limitations in conventional statistical approaches. However, current methods typically involve cluttered probability distribution function (PDF), have lower sensitivity in censoring prediction, only model static datasets, or only rely on recurrent neural networks for dynamic modelling. In this paper, we propose a novel survival regression method capable of producing high-quality unimodal PDFs without any prior distribution assumption, by optimizing novel Margin-Mean-Variance loss and leveraging the flexibility of Transformer to handle both temporal and non-temporal data, coined UniSurv. Extensive experiments on several datasets demonstrate that UniSurv places a significantly higher emphasis on censoring compared to other methods.
Generalizing CLIP to Unseen Domain via Text-Guided Diverse Novel Feature Synthesis
May 04, 2024Abstract:Vision-language foundation models like CLIP have shown impressive zero-shot generalization, but finetuning on downstream datasets can cause overfitting and loss of its generalization ability on unseen domains. Although collecting additional data from new domains of interest is possible, this method is often impractical due to the challenges in obtaining annotated data. To address this, we propose a plug-and-play feature augmentation method called LDFS (Language-Guided Diverse Feature Synthesis) to synthesize new domain features and improve existing CLIP fine-tuning strategies. LDFS has three main contributions: 1) To synthesize novel domain features and promote diversity, we propose an instance-conditional feature augmentation strategy based on a textguided feature augmentation loss. 2) To maintain feature quality after augmenting, we introduce a pairwise regularizer to preserve augmented feature coherence within the CLIP feature space. 3) We propose to use stochastic text feature augmentation to reduce the modality gap and further facilitate the process of text-guided feature synthesis. Extensive experiments show LDFS superiority in improving CLIP generalization ability on unseen domains without collecting data from those domains. The code will be made publicly available.
Progressive trajectory matching for medical dataset distillation
Mar 20, 2024Abstract:It is essential but challenging to share medical image datasets due to privacy issues, which prohibit building foundation models and knowledge transfer. In this paper, we propose a novel dataset distillation method to condense the original medical image datasets into a synthetic one that preserves useful information for building an analysis model without accessing the original datasets. Existing methods tackle only natural images by randomly matching parts of the training trajectories of the model parameters trained by the whole real datasets. However, through extensive experiments on medical image datasets, the training process is extremely unstable and achieves inferior distillation results. To solve these barriers, we propose to design a novel progressive trajectory matching strategy to improve the training stability for medical image dataset distillation. Additionally, it is observed that improved stability prevents the synthetic dataset diversity and final performance improvements. Therefore, we propose a dynamic overlap mitigation module that improves the synthetic dataset diversity by dynamically eliminating the overlap across different images and retraining parts of the synthetic images for better convergence. Finally, we propose a new medical image dataset distillation benchmark of various modalities and configurations to promote fair evaluations. It is validated that our proposed method achieves 8.33% improvement over previous state-of-the-art methods on average, and 11.7% improvement when ipc=2 (i.e., image per class is 2). Codes and benchmarks will be released.
Prompt-driven Latent Domain Generalization for Medical Image Classification
Jan 05, 2024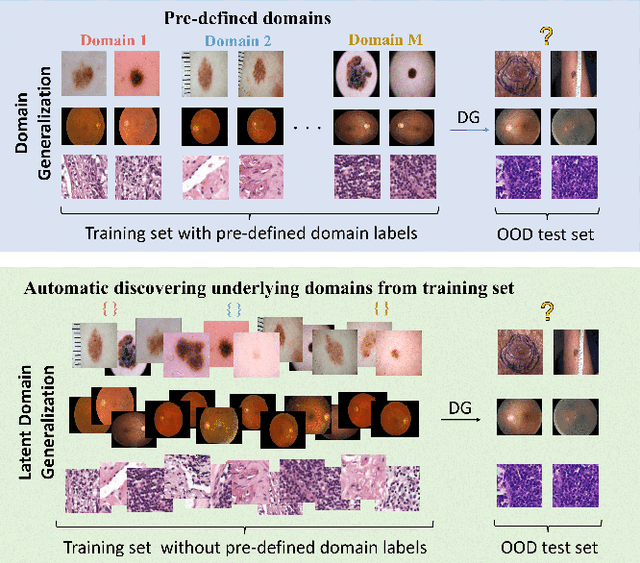
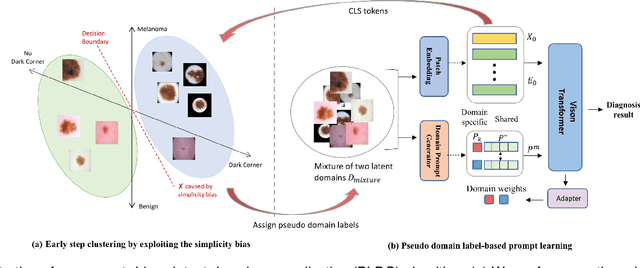
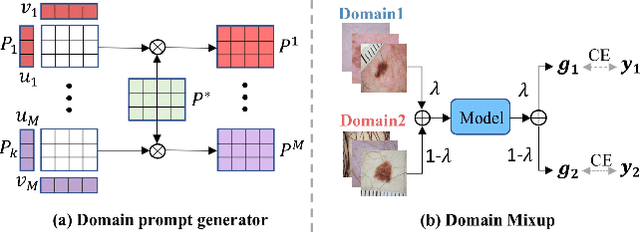
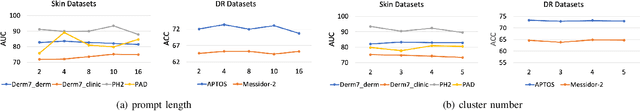
Abstract:Deep learning models for medical image analysis easily suffer from distribution shifts caused by dataset artifacts bias, camera variations, differences in the imaging station, etc., leading to unreliable diagnoses in real-world clinical settings. Domain generalization (DG) methods, which aim to train models on multiple domains to perform well on unseen domains, offer a promising direction to solve the problem. However, existing DG methods assume domain labels of each image are available and accurate, which is typically feasible for only a limited number of medical datasets. To address these challenges, we propose a novel DG framework for medical image classification without relying on domain labels, called Prompt-driven Latent Domain Generalization (PLDG). PLDG consists of unsupervised domain discovery and prompt learning. This framework first discovers pseudo domain labels by clustering the bias-associated style features, then leverages collaborative domain prompts to guide a Vision Transformer to learn knowledge from discovered diverse domains. To facilitate cross-domain knowledge learning between different prompts, we introduce a domain prompt generator that enables knowledge sharing between domain prompts and a shared prompt. A domain mixup strategy is additionally employed for more flexible decision margins and mitigates the risk of incorrect domain assignments. Extensive experiments on three medical image classification tasks and one debiasing task demonstrate that our method can achieve comparable or even superior performance than conventional DG algorithms without relying on domain labels. Our code will be publicly available upon the paper is accepted.
EPVT: Environment-aware Prompt Vision Transformer for Domain Generalization in Skin Lesion Recognition
Apr 09, 2023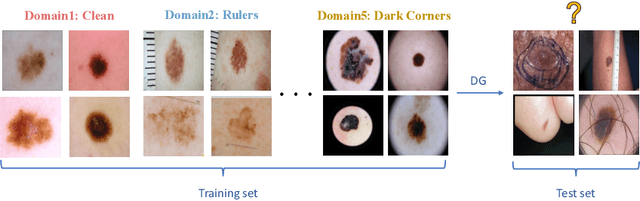
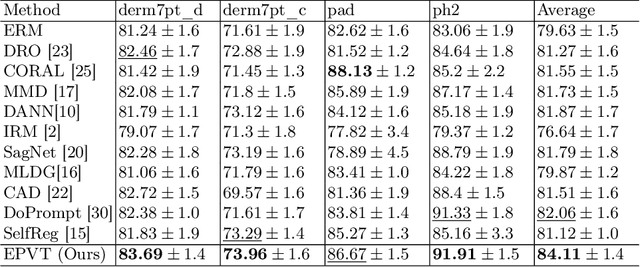
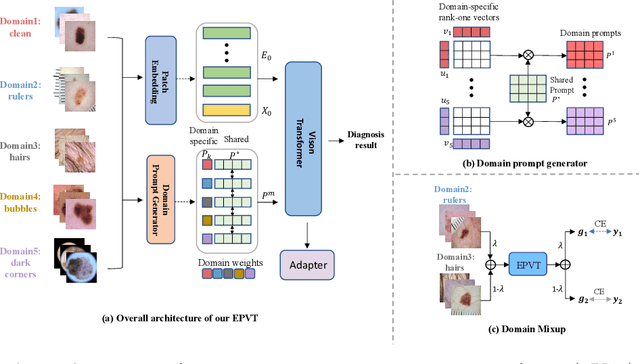
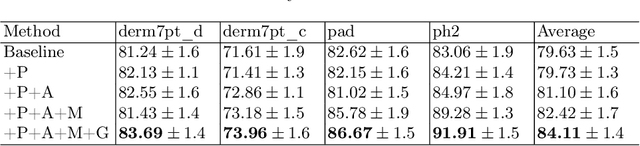
Abstract:Skin lesion recognition using deep learning has made remarkable progress, and there is an increasing need for deploying these systems in real-world scenarios. However, recent research has revealed that deep neural networks for skin lesion recognition may overly depend on disease-irrelevant image artifacts (i.e. dark corners, dense hairs), leading to poor generalization in unseen environments. To address this issue, we propose a novel domain generalization method called EPVT, which involves embedding prompts into the vision transformer to collaboratively learn knowledge from diverse domains. Concretely, EPVT leverages a set of domain prompts, each of which plays as a domain expert, to capture domain-specific knowledge; and a shared prompt for general knowledge over the entire dataset. To facilitate knowledge sharing and the interaction of different prompts, we introduce a domain prompt generator that enables low-rank multiplicative updates between domain prompts and the shared prompt. A domain mixup strategy is additionally devised to reduce the co-occurring artifacts in each domain, which allows for more flexible decision margins and mitigates the issue of incorrectly assigned domain labels. Experiments on four out-of-distribution datasets and six different biased ISIC datasets demonstrate the superior generalization ability of EPVT in skin lesion recognition across various environments. Our code and dataset will be released at https://github.com/SiyuanYan1/EPVT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge