scDD: Latent Codes Based scRNA-seq Dataset Distillation with Foundation Model Knowledge
Paper and Code
Mar 06, 2025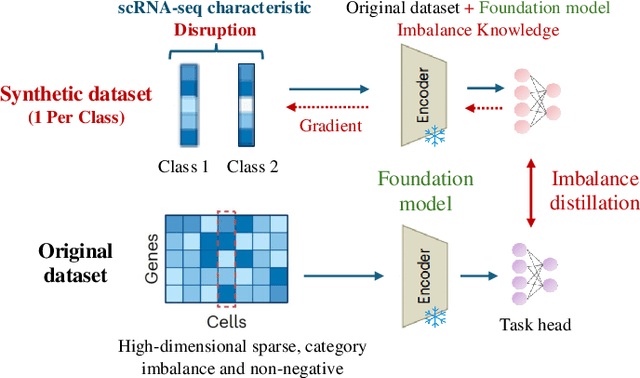
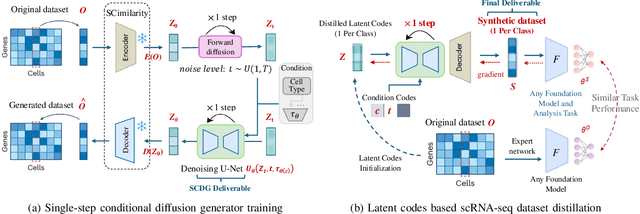
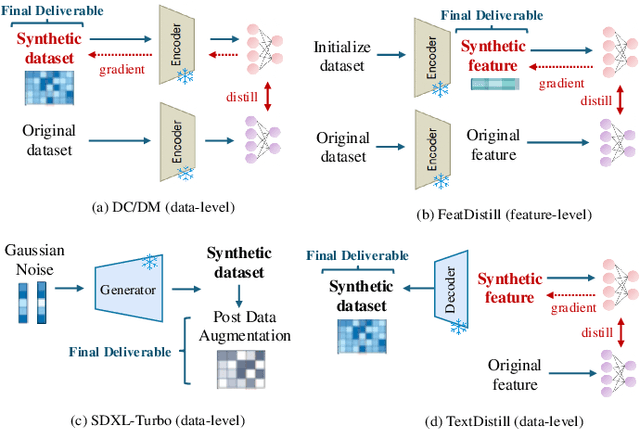
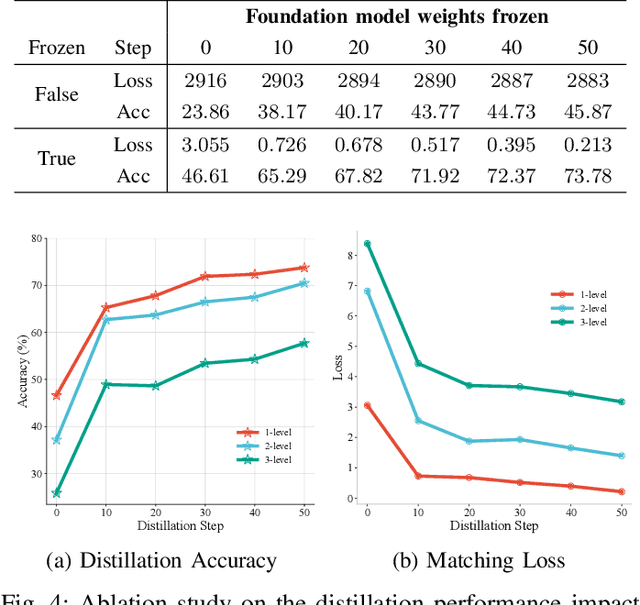
Single-cell RNA sequencing (scRNA-seq) technology has profiled hundreds of millions of human cells across organs, diseases, development and perturbations to date. However, the high-dimensional sparsity, batch effect noise, category imbalance, and ever-increasing data scale of the original sequencing data pose significant challenges for multi-center knowledge transfer, data fusion, and cross-validation between scRNA-seq datasets. To address these barriers, (1) we first propose a latent codes-based scRNA-seq dataset distillation framework named scDD, which transfers and distills foundation model knowledge and original dataset information into a compact latent space and generates synthetic scRNA-seq dataset by a generator to replace the original dataset. Then, (2) we propose a single-step conditional diffusion generator named SCDG, which perform single-step gradient back-propagation to help scDD optimize distillation quality and avoid gradient decay caused by multi-step back-propagation. Meanwhile, SCDG ensures the scRNA-seq data characteristics and inter-class discriminability of the synthetic dataset through flexible conditional control and generation quality assurance. Finally, we propose a comprehensive benchmark to evaluate the performance of scRNA-seq dataset distillation in different data analysis tasks. It is validated that our proposed method can achieve 7.61% absolute and 15.70% relative improvement over previous state-of-the-art methods on average task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge