Jianan Han
scDD: Latent Codes Based scRNA-seq Dataset Distillation with Foundation Model Knowledge
Mar 06, 2025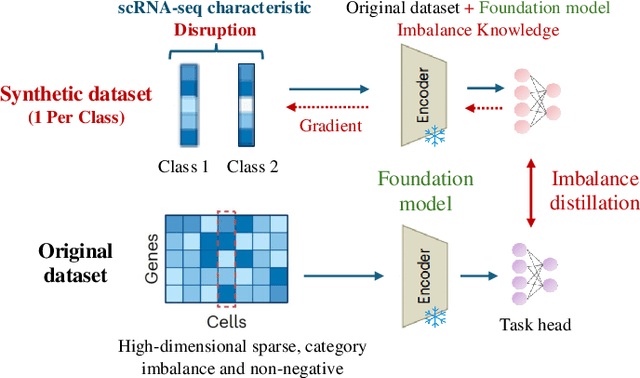
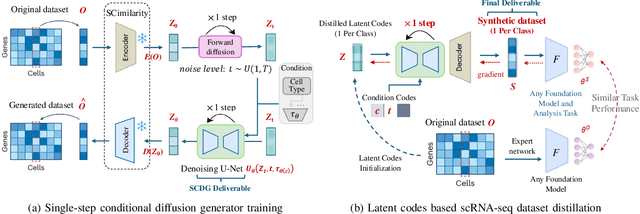
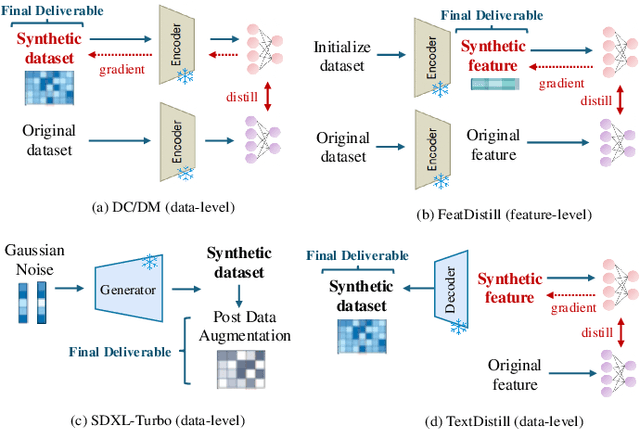
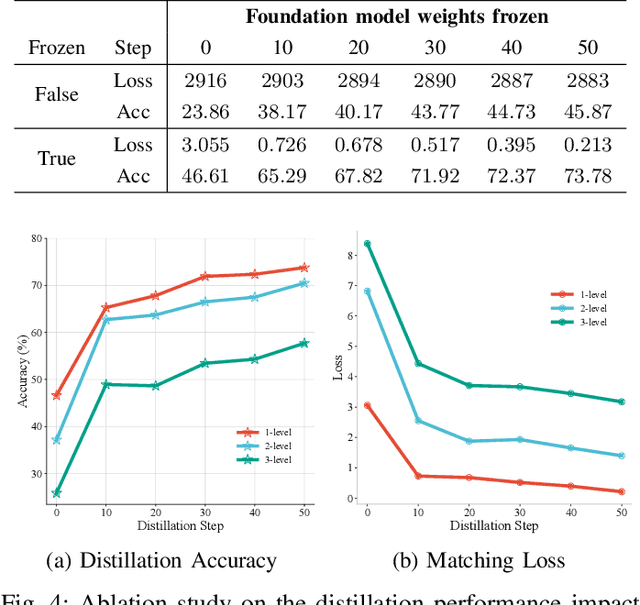
Abstract:Single-cell RNA sequencing (scRNA-seq) technology has profiled hundreds of millions of human cells across organs, diseases, development and perturbations to date. However, the high-dimensional sparsity, batch effect noise, category imbalance, and ever-increasing data scale of the original sequencing data pose significant challenges for multi-center knowledge transfer, data fusion, and cross-validation between scRNA-seq datasets. To address these barriers, (1) we first propose a latent codes-based scRNA-seq dataset distillation framework named scDD, which transfers and distills foundation model knowledge and original dataset information into a compact latent space and generates synthetic scRNA-seq dataset by a generator to replace the original dataset. Then, (2) we propose a single-step conditional diffusion generator named SCDG, which perform single-step gradient back-propagation to help scDD optimize distillation quality and avoid gradient decay caused by multi-step back-propagation. Meanwhile, SCDG ensures the scRNA-seq data characteristics and inter-class discriminability of the synthetic dataset through flexible conditional control and generation quality assurance. Finally, we propose a comprehensive benchmark to evaluate the performance of scRNA-seq dataset distillation in different data analysis tasks. It is validated that our proposed method can achieve 7.61% absolute and 15.70% relative improvement over previous state-of-the-art methods on average task.
Seeing your sleep stage: cross-modal distillation from EEG to infrared video
Aug 11, 2022



Abstract:It is inevitably crucial to classify sleep stage for the diagnosis of various diseases. However, existing automated diagnosis methods mostly adopt the "gold-standard" lectroencephalogram (EEG) or other uni-modal sensing signal of the PolySomnoGraphy (PSG) machine in hospital, that are expensive, importable and therefore unsuitable for point-of-care monitoring at home. To enable the sleep stage monitoring at home, in this paper, we analyze the relationship between infrared videos and the EEG signal and propose a new task: to classify the sleep stage using infrared videos by distilling useful knowledge from EEG signals to the visual ones. To establish a solid cross-modal benchmark for this application, we develop a new dataset termed as Seeing your Sleep Stage via Infrared Video and EEG ($S^3VE$). $S^3VE$ is a large-scale dataset including synchronized infrared video and EEG signal for sleep stage classification, including 105 subjects and 154,573 video clips that is more than 1100 hours long. Our contributions are not limited to datasets but also about a novel cross-modal distillation baseline model namely the structure-aware contrastive distillation (SACD) to distill the EEG knowledge to infrared video features. The SACD achieved the state-of-the-art performances on both our $S^3VE$ and the existing cross-modal distillation benchmark. Both the benchmark and the baseline methods will be released to the community. We expect to raise more attentions and promote more developments in the sleep stage classification and more importantly the cross-modal distillation from clinical signal/media to the conventional media.
Taylor saves for later: disentanglement for video prediction using Taylor representation
May 24, 2021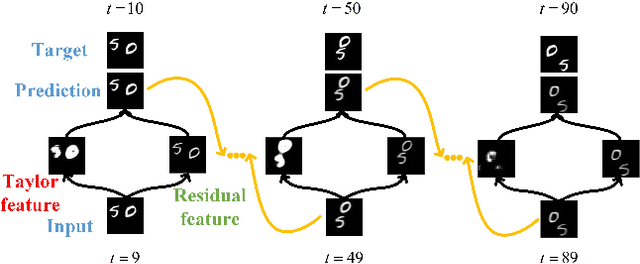

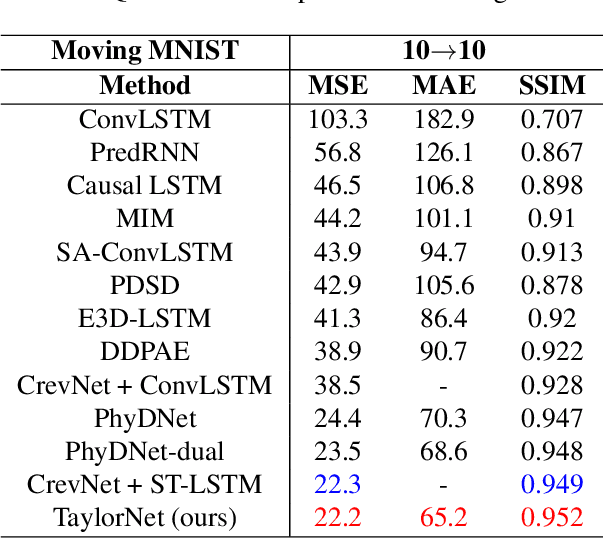
Abstract:Video prediction is a challenging task with wide application prospects in meteorology and robot systems. Existing works fail to trade off short-term and long-term prediction performances and extract robust latent dynamics laws in video frames. We propose a two-branch seq-to-seq deep model to disentangle the Taylor feature and the residual feature in video frames by a novel recurrent prediction module (TaylorCell) and residual module. TaylorCell can expand the video frames' high-dimensional features into the finite Taylor series to describe the latent laws. In TaylorCell, we propose the Taylor prediction unit (TPU) and the memory correction unit (MCU). TPU employs the first input frame's derivative information to predict the future frames, avoiding error accumulation. MCU distills all past frames' information to correct the predicted Taylor feature from TPU. Correspondingly, the residual module extracts the residual feature complementary to the Taylor feature. On three generalist datasets (Moving MNIST, TaxiBJ, Human 3.6), our model outperforms or reaches state-of-the-art models, and ablation experiments demonstrate the effectiveness of our model in long-term prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge