Dwarikanath Mahapatra
T3: Test-Time Model Merging in VLMs for Zero-Shot Medical Imaging Analysis
Oct 31, 2025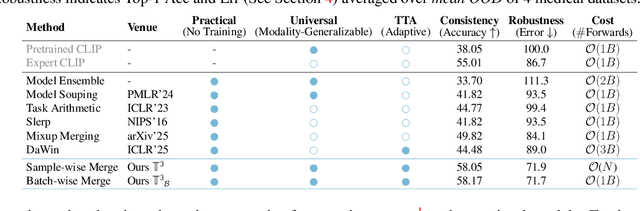
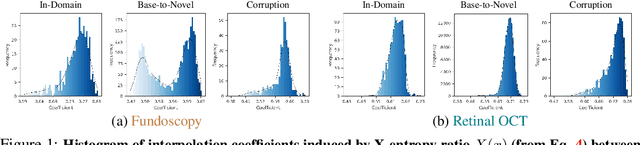
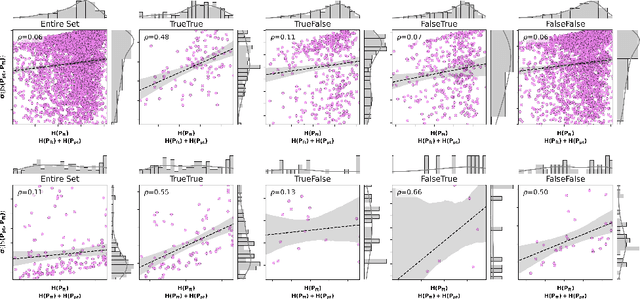
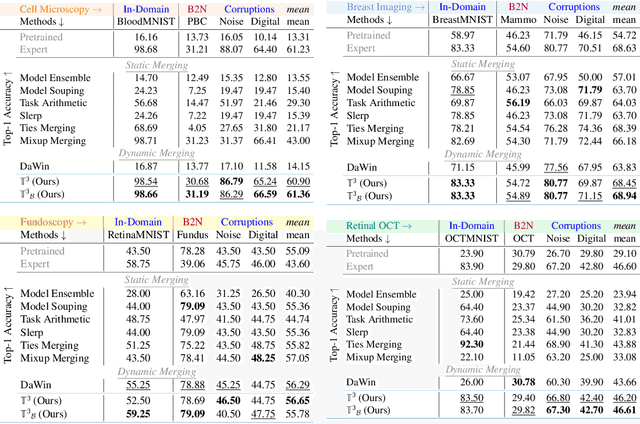
Abstract:In medical imaging, vision-language models face a critical duality: pretrained networks offer broad robustness but lack subtle, modality-specific characteristics, while fine-tuned expert models achieve high in-distribution accuracy yet falter under modality shift. Existing model-merging techniques, designed for natural-image benchmarks, are simple and efficient but fail to deliver consistent gains across diverse medical modalities; their static interpolation limits reliability in varied clinical tasks. To address this, we introduce Test-Time Task adaptive merging (T^3), a backpropagation-free framework that computes per-sample interpolation coefficients via the Jensen-Shannon divergence between the two models' output distributions. T^3 dynamically preserves local precision when models agree and defers to generalist robustness under drift. To overcome the inference costs of sample-wise merging, we further propose a batch-wise extension, T^3_B, that computes a merging coefficient across a batch of samples, dramatically reducing computational bottleneck. Recognizing the lack of a standardized medical-merging benchmark, we present a rigorous cross-evaluation protocol spanning in-domain, base-to-novel, and corruptions across four modalities. Empirically, T^3 sets new state-of-the-art in Top-1 accuracy and error reduction, outperforming strong baselines while maintaining efficiency, paving the way for adaptive MVLM deployment in clinical settings. Our code is available at https://github.com/Razaimam45/TCube.
Decoupling Clinical and Class-Agnostic Features for Reliable Few-Shot Adaptation under Shift
Sep 11, 2025Abstract:Medical vision-language models (VLMs) offer promise for clinical decision support, yet their reliability under distribution shifts remains a major concern for safe deployment. These models often learn task-agnostic correlations due to variability in imaging protocols and free-text reports, limiting their generalizability and increasing the risk of failure in real-world settings. We propose DRiFt, a structured feature decoupling framework that explicitly separates clinically relevant signals from task-agnostic noise using parameter-efficient tuning (LoRA) and learnable prompt tokens. To enhance cross-modal alignment and reduce uncertainty, we curate high-quality, clinically grounded image-text pairs by generating captions for a diverse medical dataset. Our approach improves in-distribution performance by +11.4% Top-1 accuracy and +3.3% Macro-F1 over prior prompt-based methods, while maintaining strong robustness across unseen datasets. Ablation studies reveal that disentangling task-relevant features and careful alignment significantly enhance model generalization and reduce unpredictable behavior under domain shift. These insights contribute toward building safer, more trustworthy VLMs for clinical use. The code is available at https://github.com/rumaima/DRiFt.
MuGa-VTON: Multi-Garment Virtual Try-On via Diffusion Transformers with Prompt Customization
Aug 11, 2025Abstract:Virtual try-on seeks to generate photorealistic images of individuals in desired garments, a task that must simultaneously preserve personal identity and garment fidelity for practical use in fashion retail and personalization. However, existing methods typically handle upper and lower garments separately, rely on heavy preprocessing, and often fail to preserve person-specific cues such as tattoos, accessories, and body shape-resulting in limited realism and flexibility. To this end, we introduce MuGa-VTON, a unified multi-garment diffusion framework that jointly models upper and lower garments together with person identity in a shared latent space. Specifically, we proposed three key modules: the Garment Representation Module (GRM) for capturing both garment semantics, the Person Representation Module (PRM) for encoding identity and pose cues, and the A-DiT fusion module, which integrates garment, person, and text-prompt features through a diffusion transformer. This architecture supports prompt-based customization, allowing fine-grained garment modifications with minimal user input. Extensive experiments on the VITON-HD and DressCode benchmarks demonstrate that MuGa-VTON outperforms existing methods in both qualitative and quantitative evaluations, producing high-fidelity, identity-preserving results suitable for real-world virtual try-on applications.
Proceedings of 1st Workshop on Advancing Artificial Intelligence through Theory of Mind
Apr 28, 2025

Abstract:This volume includes a selection of papers presented at the Workshop on Advancing Artificial Intelligence through Theory of Mind held at AAAI 2025 in Philadelphia US on 3rd March 2025. The purpose of this volume is to provide an open access and curated anthology for the ToM and AI research community.
Right Prediction, Wrong Reasoning: Uncovering LLM Misalignment in RA Disease Diagnosis
Apr 09, 2025Abstract:Large language models (LLMs) offer a promising pre-screening tool, improving early disease detection and providing enhanced healthcare access for underprivileged communities. The early diagnosis of various diseases continues to be a significant challenge in healthcare, primarily due to the nonspecific nature of early symptoms, the shortage of expert medical practitioners, and the need for prolonged clinical evaluations, all of which can delay treatment and adversely affect patient outcomes. With impressive accuracy in prediction across a range of diseases, LLMs have the potential to revolutionize clinical pre-screening and decision-making for various medical conditions. In this work, we study the diagnostic capability of LLMs for Rheumatoid Arthritis (RA) with real world patients data. Patient data was collected alongside diagnoses from medical experts, and the performance of LLMs was evaluated in comparison to expert diagnoses for RA disease prediction. We notice an interesting pattern in disease diagnosis and find an unexpected \textit{misalignment between prediction and explanation}. We conduct a series of multi-round analyses using different LLM agents. The best-performing model accurately predicts rheumatoid arthritis (RA) diseases approximately 95\% of the time. However, when medical experts evaluated the reasoning generated by the model, they found that nearly 68\% of the reasoning was incorrect. This study highlights a clear misalignment between LLMs high prediction accuracy and its flawed reasoning, raising important questions about relying on LLM explanations in clinical settings. \textbf{LLMs provide incorrect reasoning to arrive at the correct answer for RA disease diagnosis.}
Slide-Level Prompt Learning with Vision Language Models for Few-Shot Multiple Instance Learning in Histopathology
Mar 21, 2025Abstract:In this paper, we address the challenge of few-shot classification in histopathology whole slide images (WSIs) by utilizing foundational vision-language models (VLMs) and slide-level prompt learning. Given the gigapixel scale of WSIs, conventional multiple instance learning (MIL) methods rely on aggregation functions to derive slide-level (bag-level) predictions from patch representations, which require extensive bag-level labels for training. In contrast, VLM-based approaches excel at aligning visual embeddings of patches with candidate class text prompts but lack essential pathological prior knowledge. Our method distinguishes itself by utilizing pathological prior knowledge from language models to identify crucial local tissue types (patches) for WSI classification, integrating this within a VLM-based MIL framework. Our approach effectively aligns patch images with tissue types, and we fine-tune our model via prompt learning using only a few labeled WSIs per category. Experimentation on real-world pathological WSI datasets and ablation studies highlight our method's superior performance over existing MIL- and VLM-based methods in few-shot WSI classification tasks. Our code is publicly available at https://github.com/LTS5/SLIP.
Gene42: Long-Range Genomic Foundation Model With Dense Attention
Mar 20, 2025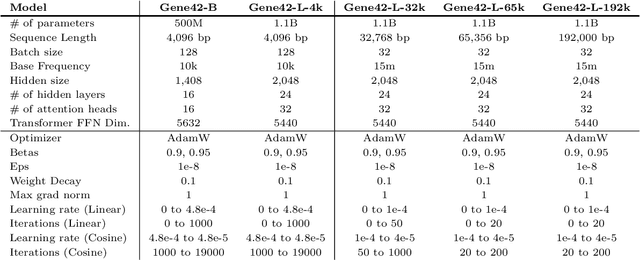
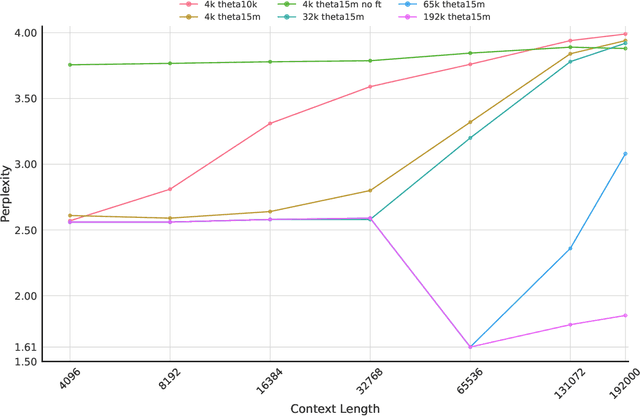
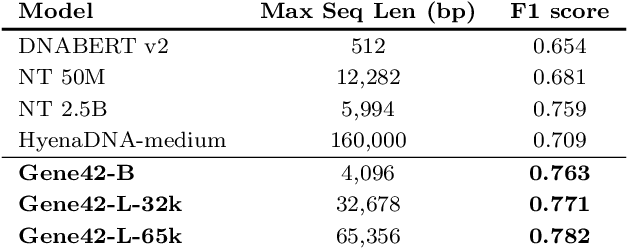
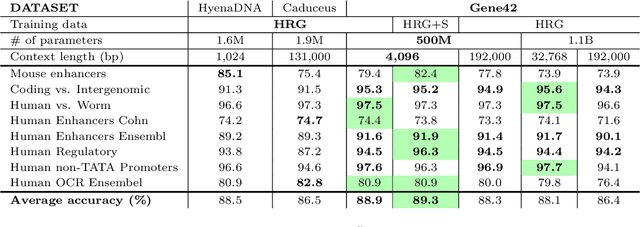
Abstract:We introduce Gene42, a novel family of Genomic Foundation Models (GFMs) designed to manage context lengths of up to 192,000 base pairs (bp) at a single-nucleotide resolution. Gene42 models utilize a decoder-only (LLaMA-style) architecture with a dense self-attention mechanism. Initially trained on fixed-length sequences of 4,096 bp, our models underwent continuous pretraining to extend the context length to 192,000 bp. This iterative extension allowed for the comprehensive processing of large-scale genomic data and the capture of intricate patterns and dependencies within the human genome. Gene42 is the first dense attention model capable of handling such extensive long context lengths in genomics, challenging state-space models that often rely on convolutional operators among other mechanisms. Our pretrained models exhibit notably low perplexity values and high reconstruction accuracy, highlighting their strong ability to model genomic data. Extensive experiments on various genomic benchmarks have demonstrated state-of-the-art performance across multiple tasks, including biotype classification, regulatory region identification, chromatin profiling prediction, variant pathogenicity prediction, and species classification. The models are publicly available at huggingface.co/inceptionai.
MedUnA: Language guided Unsupervised Adaptation of Vision-Language Models for Medical Image Classification
Sep 03, 2024Abstract:In medical image classification, supervised learning is challenging due to the lack of labeled medical images. Contrary to the traditional \textit{modus operandi} of pre-training followed by fine-tuning, this work leverages the visual-textual alignment within Vision-Language models (\texttt{VLMs}) to facilitate the unsupervised learning. Specifically, we propose \underline{Med}ical \underline{Un}supervised \underline{A}daptation (\texttt{MedUnA}), constituting two-stage training: Adapter Pre-training, and Unsupervised Learning. In the first stage, we use descriptions generated by a Large Language Model (\texttt{LLM}) corresponding to class labels, which are passed through the text encoder \texttt{BioBERT}. The resulting text embeddings are then aligned with the class labels by training a lightweight \texttt{adapter}. We choose \texttt{\texttt{LLMs}} because of their capability to generate detailed, contextually relevant descriptions to obtain enhanced text embeddings. In the second stage, the trained \texttt{adapter} is integrated with the visual encoder of \texttt{MedCLIP}. This stage employs a contrastive entropy-based loss and prompt tuning to align visual embeddings. We incorporate self-entropy minimization into the overall training objective to ensure more confident embeddings, which are crucial for effective unsupervised learning and alignment. We evaluate the performance of \texttt{MedUnA} on three different kinds of data modalities - chest X-rays, eye fundus and skin lesion images. The results demonstrate significant accuracy gain on average compared to the baselines across different datasets, highlighting the efficacy of our approach.
Characterizing Continual Learning Scenarios and Strategies for Audio Analysis
Jun 29, 2024



Abstract:Audio analysis is useful in many application scenarios. The state-of-the-art audio analysis approaches assume that the data distribution at training and deployment time will be the same. However, due to various real-life environmental factors, the data may encounter drift in its distribution or can encounter new classes in the late future. Thus, a one-time trained model might not perform adequately. In this paper, we characterize continual learning (CL) approaches in audio analysis. In this paper, we characterize continual learning (CL) approaches, intended to tackle catastrophic forgetting arising due to drifts. As there is no CL dataset for audio analysis, we use DCASE 2020 to 2023 datasets to create various CL scenarios for audio-based monitoring tasks. We have investigated the following CL and non-CL approaches: EWC, LwF, SI, GEM, A-GEM, GDumb, Replay, Naive, cumulative, and joint training. The study is very beneficial for researchers and practitioners working in the area of audio analysis for developing adaptive models. We observed that Replay achieved better results than other methods in the DCASE challenge data. It achieved an accuracy of 70.12% for the domain incremental scenario and an accuracy of 96.98% for the class incremental scenario.
Envisioning MedCLIP: A Deep Dive into Explainability for Medical Vision-Language Models
Mar 27, 2024Abstract:Explaining Deep Learning models is becoming increasingly important in the face of daily emerging multimodal models, particularly in safety-critical domains like medical imaging. However, the lack of detailed investigations into the performance of explainability methods on these models is widening the gap between their development and safe deployment. In this work, we analyze the performance of various explainable AI methods on a vision-language model, MedCLIP, to demystify its inner workings. We also provide a simple methodology to overcome the shortcomings of these methods. Our work offers a different new perspective on the explainability of a recent well-known VLM in the medical domain and our assessment method is generalizable to other current and possible future VLMs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge