Boulbaba Ben Amor
BYOL: Bring Your Own Language Into LLMs
Jan 15, 2026Abstract:Large Language Models (LLMs) exhibit strong multilingual capabilities, yet remain fundamentally constrained by the severe imbalance in global language resources. While over 7,000 languages are spoken worldwide, only a small subset (fewer than 100) has sufficient digital presence to meaningfully influence modern LLM training. This disparity leads to systematic underperformance, cultural misalignment, and limited accessibility for speakers of low-resource and extreme-low-resource languages. To address this gap, we introduce Bring Your Own Language (BYOL), a unified framework for scalable, language-aware LLM development tailored to each language's digital footprint. BYOL begins with a language resource classification that maps languages into four tiers (Extreme-Low, Low, Mid, High) using curated web-scale corpora, and uses this classification to select the appropriate integration pathway. For low-resource languages, we propose a full-stack data refinement and expansion pipeline that combines corpus cleaning, synthetic text generation, continual pretraining, and supervised finetuning. Applied to Chichewa and Maori, this pipeline yields language-specific LLMs that achieve approximately 12 percent average improvement over strong multilingual baselines across 12 benchmarks, while preserving English and multilingual capabilities via weight-space model merging. For extreme-low-resource languages, we introduce a translation-mediated inclusion pathway, and show on Inuktitut that a tailored machine translation system improves over a commercial baseline by 4 BLEU, enabling high-accuracy LLM access when direct language modeling is infeasible. Finally, we release human-translated versions of the Global MMLU-Lite benchmark in Chichewa, Maori, and Inuktitut, and make our codebase and models publicly available at https://github.com/microsoft/byol .
Prot42: a Novel Family of Protein Language Models for Target-aware Protein Binder Generation
Apr 06, 2025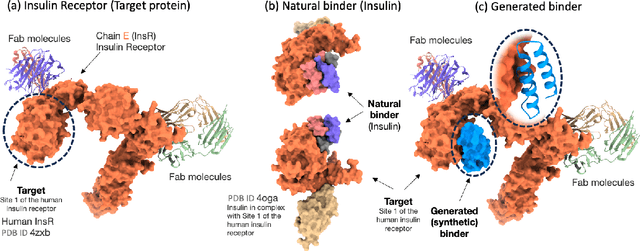
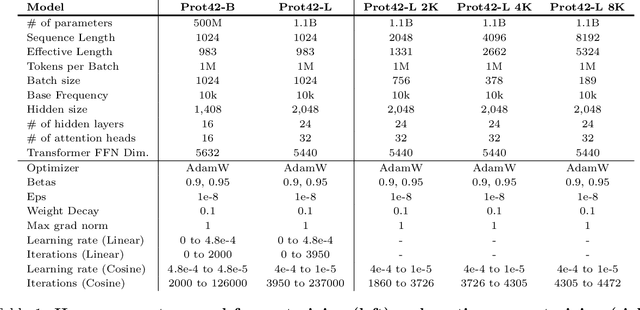
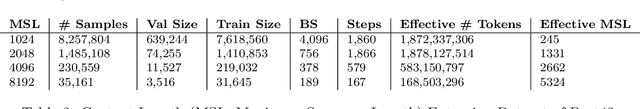
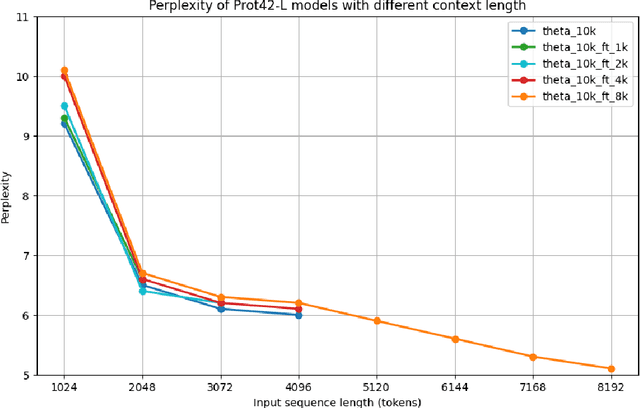
Abstract:Unlocking the next generation of biotechnology and therapeutic innovation demands overcoming the inherent complexity and resource-intensity of conventional protein engineering methods. Recent GenAI-powered computational techniques often rely on the availability of the target protein's 3D structures and specific binding sites to generate high-affinity binders, constraints exhibited by models such as AlphaProteo and RFdiffusion. In this work, we explore the use of Protein Language Models (pLMs) for high-affinity binder generation. We introduce Prot42, a novel family of Protein Language Models (pLMs) pretrained on vast amounts of unlabeled protein sequences. By capturing deep evolutionary, structural, and functional insights through an advanced auto-regressive, decoder-only architecture inspired by breakthroughs in natural language processing, Prot42 dramatically expands the capabilities of computational protein design based on language only. Remarkably, our models handle sequences up to 8,192 amino acids, significantly surpassing standard limitations and enabling precise modeling of large proteins and complex multi-domain sequences. Demonstrating powerful practical applications, Prot42 excels in generating high-affinity protein binders and sequence-specific DNA-binding proteins. Our innovative models are publicly available, offering the scientific community an efficient and precise computational toolkit for rapid protein engineering.
Chem42: a Family of chemical Language Models for Target-aware Ligand Generation
Mar 20, 2025Abstract:Revolutionizing drug discovery demands more than just understanding molecular interactions - it requires generative models that can design novel ligands tailored to specific biological targets. While chemical Language Models (cLMs) have made strides in learning molecular properties, most fail to incorporate target-specific insights, restricting their ability to drive de-novo ligand generation. Chem42, a cutting-edge family of generative chemical Language Models, is designed to bridge this gap. By integrating atomic-level interactions with multimodal inputs from Prot42, a complementary protein Language Model, Chem42 achieves a sophisticated cross-modal representation of molecular structures, interactions, and binding patterns. This innovative framework enables the creation of structurally valid, synthetically accessible ligands with enhanced target specificity. Evaluations across diverse protein targets confirm that Chem42 surpasses existing approaches in chemical validity, target-aware design, and predicted binding affinity. By reducing the search space of viable drug candidates, Chem42 could accelerate the drug discovery pipeline, offering a powerful generative AI tool for precision medicine. Our Chem42 models set a new benchmark in molecule property prediction, conditional molecule generation, and target-aware ligand design. The models are publicly available at huggingface.co/inceptionai.
Gene42: Long-Range Genomic Foundation Model With Dense Attention
Mar 20, 2025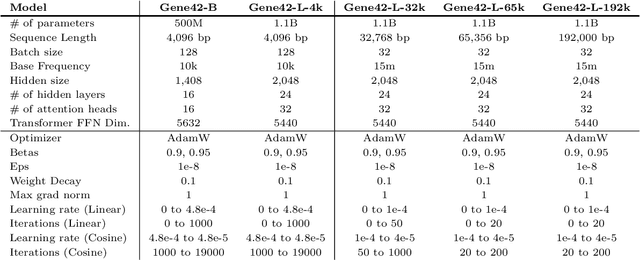
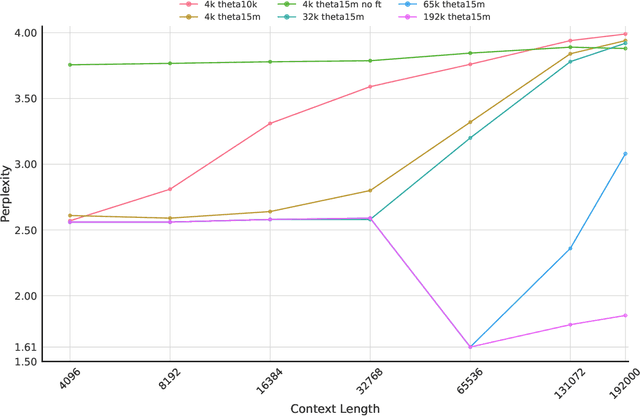
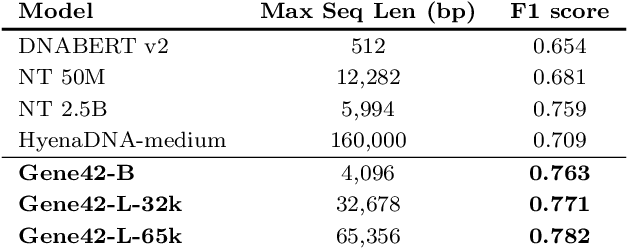
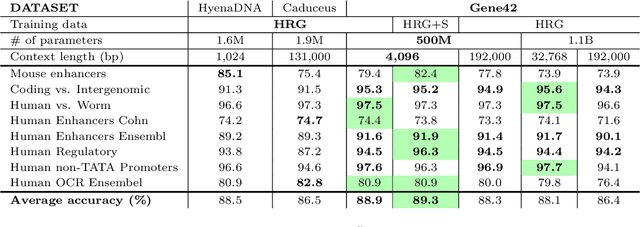
Abstract:We introduce Gene42, a novel family of Genomic Foundation Models (GFMs) designed to manage context lengths of up to 192,000 base pairs (bp) at a single-nucleotide resolution. Gene42 models utilize a decoder-only (LLaMA-style) architecture with a dense self-attention mechanism. Initially trained on fixed-length sequences of 4,096 bp, our models underwent continuous pretraining to extend the context length to 192,000 bp. This iterative extension allowed for the comprehensive processing of large-scale genomic data and the capture of intricate patterns and dependencies within the human genome. Gene42 is the first dense attention model capable of handling such extensive long context lengths in genomics, challenging state-space models that often rely on convolutional operators among other mechanisms. Our pretrained models exhibit notably low perplexity values and high reconstruction accuracy, highlighting their strong ability to model genomic data. Extensive experiments on various genomic benchmarks have demonstrated state-of-the-art performance across multiple tasks, including biotype classification, regulatory region identification, chromatin profiling prediction, variant pathogenicity prediction, and species classification. The models are publicly available at huggingface.co/inceptionai.
MedUnA: Language guided Unsupervised Adaptation of Vision-Language Models for Medical Image Classification
Sep 03, 2024Abstract:In medical image classification, supervised learning is challenging due to the lack of labeled medical images. Contrary to the traditional \textit{modus operandi} of pre-training followed by fine-tuning, this work leverages the visual-textual alignment within Vision-Language models (\texttt{VLMs}) to facilitate the unsupervised learning. Specifically, we propose \underline{Med}ical \underline{Un}supervised \underline{A}daptation (\texttt{MedUnA}), constituting two-stage training: Adapter Pre-training, and Unsupervised Learning. In the first stage, we use descriptions generated by a Large Language Model (\texttt{LLM}) corresponding to class labels, which are passed through the text encoder \texttt{BioBERT}. The resulting text embeddings are then aligned with the class labels by training a lightweight \texttt{adapter}. We choose \texttt{\texttt{LLMs}} because of their capability to generate detailed, contextually relevant descriptions to obtain enhanced text embeddings. In the second stage, the trained \texttt{adapter} is integrated with the visual encoder of \texttt{MedCLIP}. This stage employs a contrastive entropy-based loss and prompt tuning to align visual embeddings. We incorporate self-entropy minimization into the overall training objective to ensure more confident embeddings, which are crucial for effective unsupervised learning and alignment. We evaluate the performance of \texttt{MedUnA} on three different kinds of data modalities - chest X-rays, eye fundus and skin lesion images. The results demonstrate significant accuracy gain on average compared to the baselines across different datasets, highlighting the efficacy of our approach.
Med42 -- Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches
Apr 23, 2024



Abstract:This study presents a comprehensive analysis and comparison of two predominant fine-tuning methodologies - full-parameter fine-tuning and parameter-efficient tuning - within the context of medical Large Language Models (LLMs). We developed and refined a series of LLMs, based on the Llama-2 architecture, specifically designed to enhance medical knowledge retrieval, reasoning, and question-answering capabilities. Our experiments systematically evaluate the effectiveness of these tuning strategies across various well-known medical benchmarks. Notably, our medical LLM Med42 showed an accuracy level of 72% on the US Medical Licensing Examination (USMLE) datasets, setting a new standard in performance for openly available medical LLMs. Through this comparative analysis, we aim to identify the most effective and efficient method for fine-tuning LLMs in the medical domain, thereby contributing significantly to the advancement of AI-driven healthcare applications.
SHREC'22 Track: Sketch-Based 3D Shape Retrieval in the Wild
Jul 11, 2022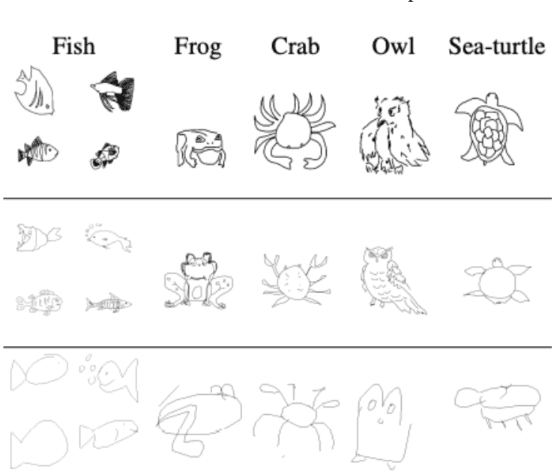
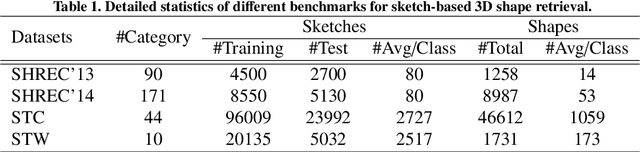
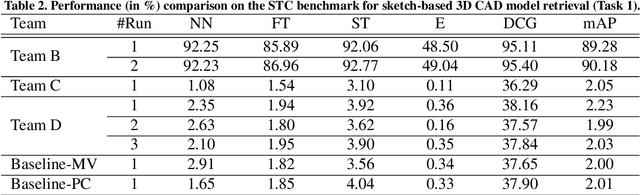
Abstract:Sketch-based 3D shape retrieval (SBSR) is an important yet challenging task, which has drawn more and more attention in recent years. Existing approaches address the problem in a restricted setting, without appropriately simulating real application scenarios. To mimic the realistic setting, in this track, we adopt large-scale sketches drawn by amateurs of different levels of drawing skills, as well as a variety of 3D shapes including not only CAD models but also models scanned from real objects. We define two SBSR tasks and construct two benchmarks consisting of more than 46,000 CAD models, 1,700 realistic models, and 145,000 sketches in total. Four teams participated in this track and submitted 15 runs for the two tasks, evaluated by 7 commonly-adopted metrics. We hope that, the benchmarks, the comparative results, and the open-sourced evaluation code will foster future research in this direction among the 3D object retrieval community.
G-VAE, a Geometric Convolutional VAE for ProteinStructure Generation
Jun 22, 2021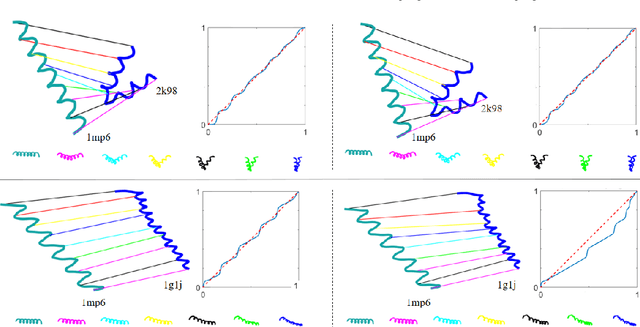
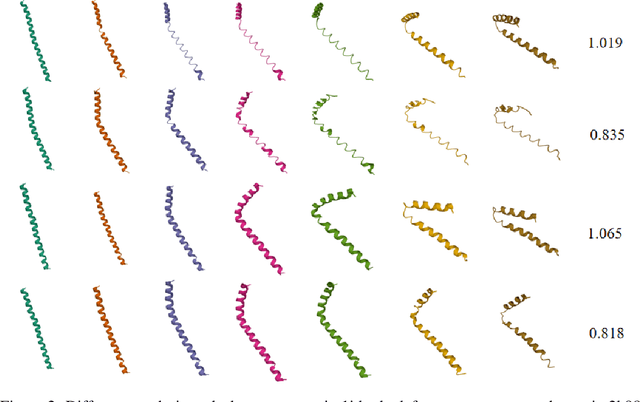
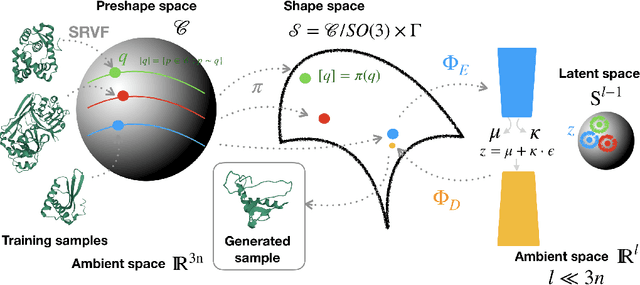
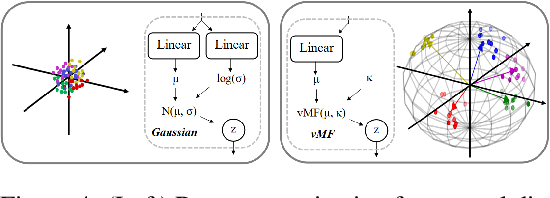
Abstract:Analyzing the structure of proteins is a key part of understanding their functions and thus their role in biology at the molecular level. In addition, design new proteins in a methodical way is a major engineering challenge. In this work, we introduce a joint geometric-neural networks approach for comparing, deforming and generating 3D protein structures. Viewing protein structures as 3D open curves, we adopt the Square Root Velocity Function (SRVF) representation and leverage its suitable geometric properties along with Deep Residual Networks (ResNets) for a joint registration and comparison. Our ResNets handle better large protein deformations while being more computationally efficient. On top of the mathematical framework, we further design a Geometric Variational Auto-Encoder (G-VAE), that once trained, maps original, previously unseen structures, into a low-dimensional (latent) hyper-sphere. Motivated by the spherical structure of the pre-shape space, we naturally adopt the von Mises-Fisher (vMF) distribution to model our hidden variables. We test the effectiveness of our models by generating novel protein structures and predicting completions of corrupted protein structures. Experimental results show that our method is able to generate plausible structures, different from the structures in the training data.
Residual Networks as Flows of Velocity Fields for Diffeomorphic Time Series Alignment
Jun 22, 2021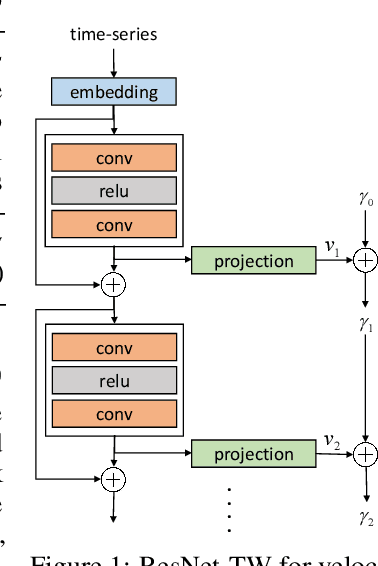
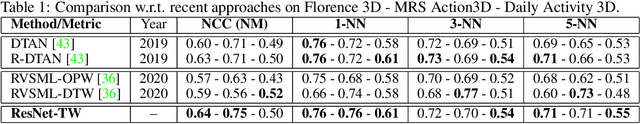
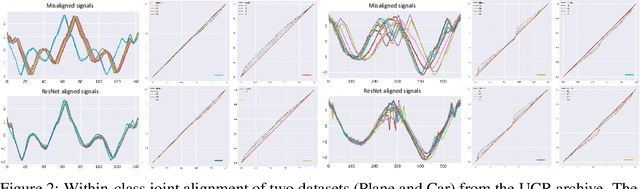
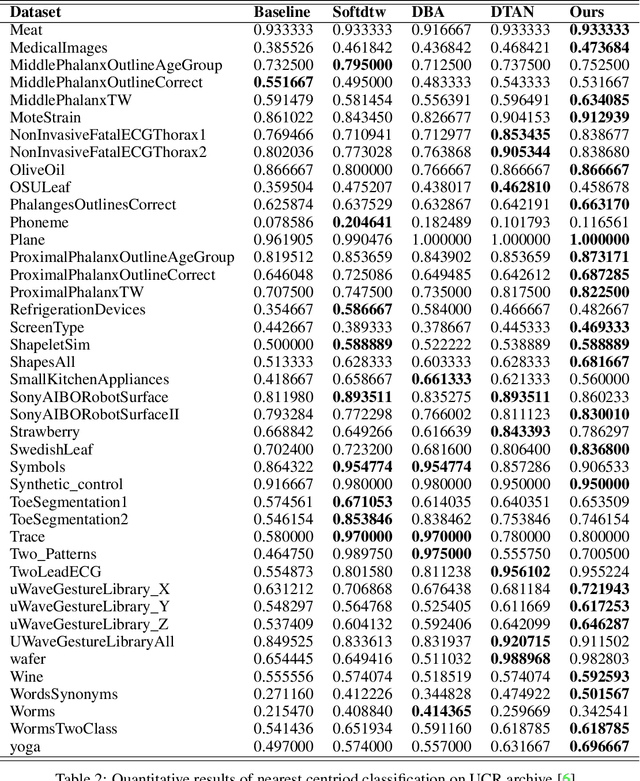
Abstract:Non-linear (large) time warping is a challenging source of nuisance in time-series analysis. In this paper, we propose a novel diffeomorphic temporal transformer network for both pairwise and joint time-series alignment. Our ResNet-TW (Deep Residual Network for Time Warping) tackles the alignment problem by compositing a flow of incremental diffeomorphic mappings. Governed by the flow equation, our Residual Network (ResNet) builds smooth, fluid and regular flows of velocity fields and consequently generates smooth and invertible transformations (i.e. diffeomorphic warping functions). Inspired by the elegant Large Deformation Diffeomorphic Metric Mapping (LDDMM) framework, the final transformation is built by the flow of time-dependent vector fields which are none other than the building blocks of our Residual Network. The latter is naturally viewed as an Eulerian discretization schema of the flow equation (an ODE). Once trained, our ResNet-TW aligns unseen data by a single inexpensive forward pass. As we show in experiments on both univariate (84 datasets from UCR archive) and multivariate time-series (MSR Action-3D, Florence-3D and MSR Daily Activity), ResNet-TW achieves competitive performance in joint alignment and classification.
Magnifying Subtle Facial Motions for Effective 4D Expression Recognition
May 05, 2021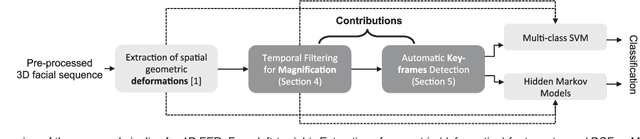
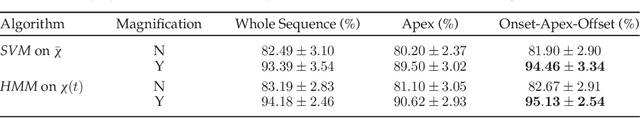
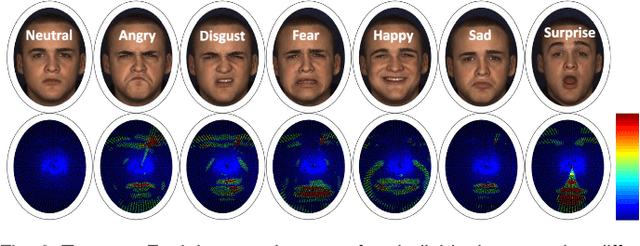
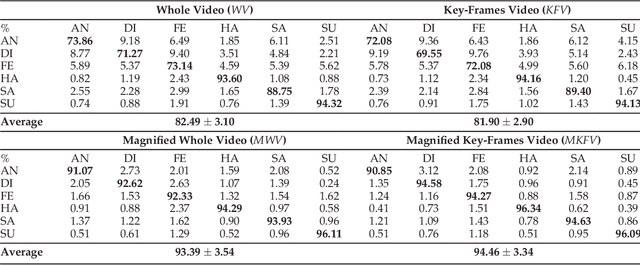
Abstract:In this paper, an effective pipeline to automatic 4D Facial Expression Recognition (4D FER) is proposed. It combines two growing but disparate ideas in Computer Vision -- computing the spatial facial deformations using tools from Riemannian geometry and magnifying them using temporal filtering. The flow of 3D faces is first analyzed to capture the spatial deformations based on the recently-developed Riemannian approach, where registration and comparison of neighboring 3D faces are led jointly. Then, the obtained temporal evolution of these deformations are fed into a magnification method in order to amplify the facial activities over the time. The latter, main contribution of this paper, allows revealing subtle (hidden) deformations which enhance the emotion classification performance. We evaluated our approach on BU-4DFE dataset, the state-of-art 94.18% average performance and an improvement that exceeds 10% in classification accuracy, after magnifying extracted geometric features (deformations), are achieved.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge