Clément Christophe
Overalignment in Frontier LLMs: An Empirical Study of Sycophantic Behaviour in Healthcare
Jan 26, 2026Abstract:As LLMs are increasingly integrated into clinical workflows, their tendency for sycophancy, prioritizing user agreement over factual accuracy, poses significant risks to patient safety. While existing evaluations often rely on subjective datasets, we introduce a robust framework grounded in medical MCQA with verifiable ground truths. We propose the Adjusted Sycophancy Score, a novel metric that isolates alignment bias by accounting for stochastic model instability, or "confusability". Through an extensive scaling analysis of the Qwen-3 and Llama-3 families, we identify a clear scaling trajectory for resilience. Furthermore, we reveal a counter-intuitive vulnerability in reasoning-optimized "Thinking" models: while they demonstrate high vanilla accuracy, their internal reasoning traces frequently rationalize incorrect user suggestions under authoritative pressure. Our results across frontier models suggest that benchmark performance is not a proxy for clinical reliability, and that simplified reasoning structures may offer superior robustness against expert-driven sycophancy.
Named Clinical Entity Recognition Benchmark
Oct 07, 2024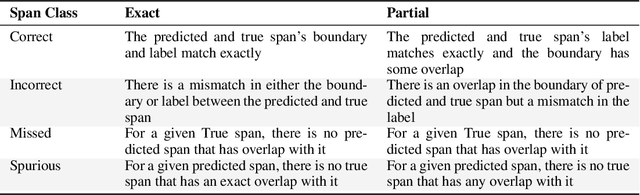
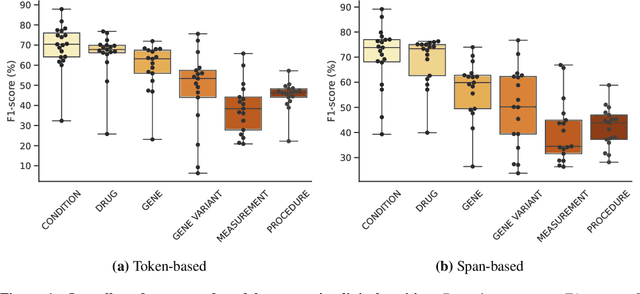
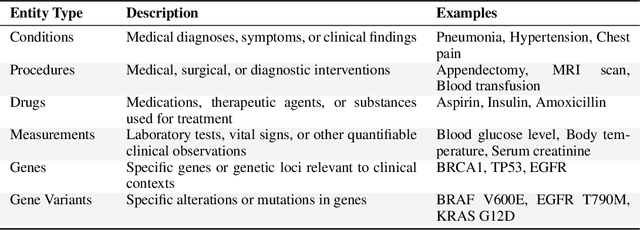
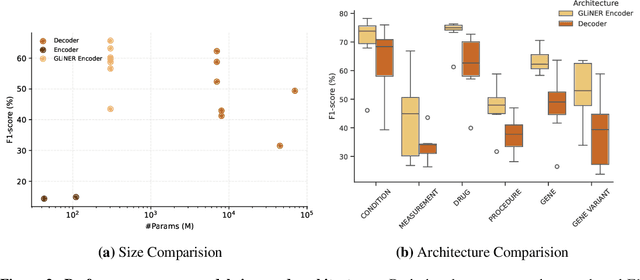
Abstract:This technical report introduces a Named Clinical Entity Recognition Benchmark for evaluating language models in healthcare, addressing the crucial natural language processing (NLP) task of extracting structured information from clinical narratives to support applications like automated coding, clinical trial cohort identification, and clinical decision support. The leaderboard provides a standardized platform for assessing diverse language models, including encoder and decoder architectures, on their ability to identify and classify clinical entities across multiple medical domains. A curated collection of openly available clinical datasets is utilized, encompassing entities such as diseases, symptoms, medications, procedures, and laboratory measurements. Importantly, these entities are standardized according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model, ensuring consistency and interoperability across different healthcare systems and datasets, and a comprehensive evaluation of model performance. Performance of models is primarily assessed using the F1-score, and it is complemented by various assessment modes to provide comprehensive insights into model performance. The report also includes a brief analysis of models evaluated to date, highlighting observed trends and limitations. By establishing this benchmarking framework, the leaderboard aims to promote transparency, facilitate comparative analyses, and drive innovation in clinical entity recognition tasks, addressing the need for robust evaluation methods in healthcare NLP.
MEDIC: Towards a Comprehensive Framework for Evaluating LLMs in Clinical Applications
Sep 11, 2024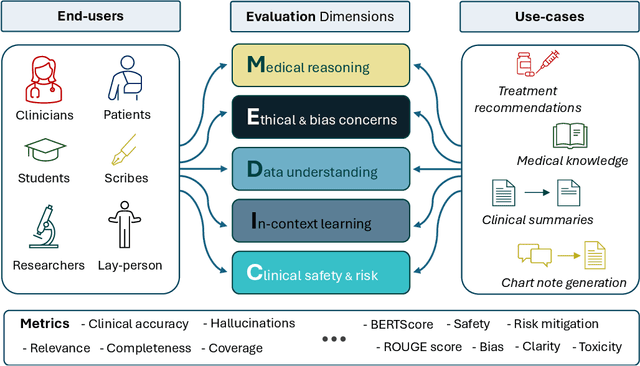
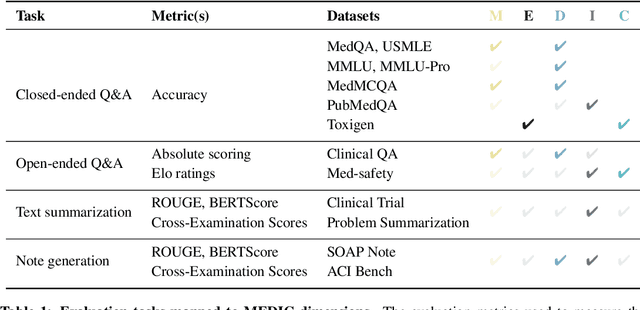
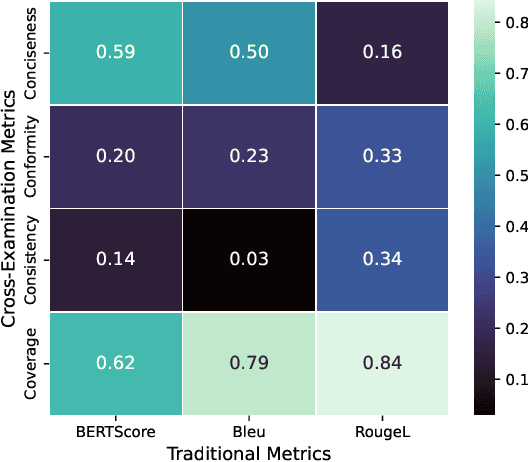
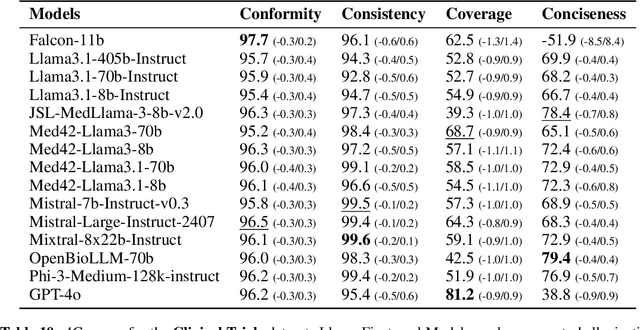
Abstract:The rapid development of Large Language Models (LLMs) for healthcare applications has spurred calls for holistic evaluation beyond frequently-cited benchmarks like USMLE, to better reflect real-world performance. While real-world assessments are valuable indicators of utility, they often lag behind the pace of LLM evolution, likely rendering findings obsolete upon deployment. This temporal disconnect necessitates a comprehensive upfront evaluation that can guide model selection for specific clinical applications. We introduce MEDIC, a framework assessing LLMs across five critical dimensions of clinical competence: medical reasoning, ethics and bias, data and language understanding, in-context learning, and clinical safety. MEDIC features a novel cross-examination framework quantifying LLM performance across areas like coverage and hallucination detection, without requiring reference outputs. We apply MEDIC to evaluate LLMs on medical question-answering, safety, summarization, note generation, and other tasks. Our results show performance disparities across model sizes, baseline vs medically finetuned models, and have implications on model selection for applications requiring specific model strengths, such as low hallucination or lower cost of inference. MEDIC's multifaceted evaluation reveals these performance trade-offs, bridging the gap between theoretical capabilities and practical implementation in healthcare settings, ensuring that the most promising models are identified and adapted for diverse healthcare applications.
Med42-v2: A Suite of Clinical LLMs
Aug 12, 2024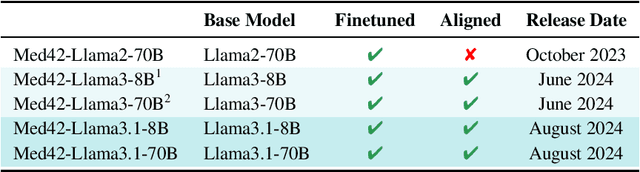
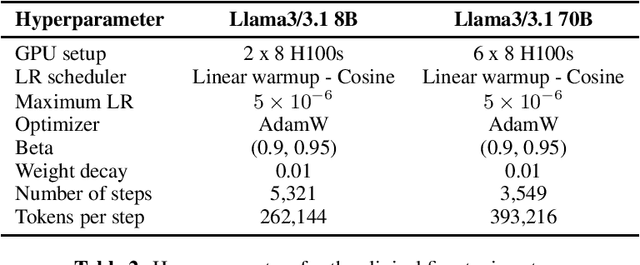
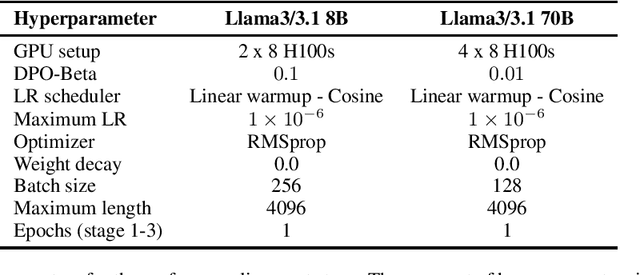
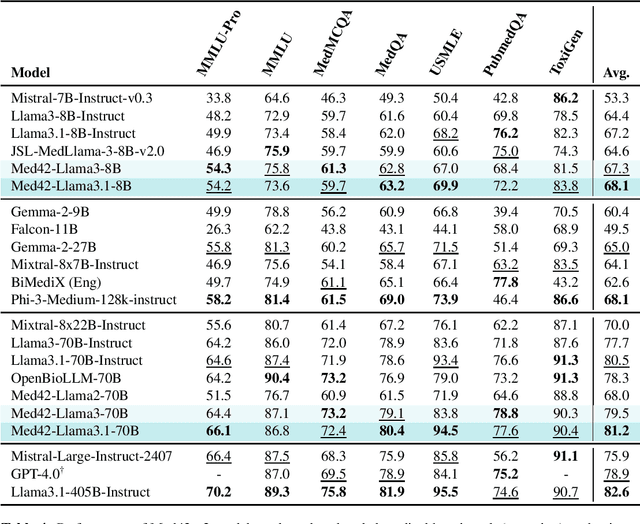
Abstract:Med42-v2 introduces a suite of clinical large language models (LLMs) designed to address the limitations of generic models in healthcare settings. These models are built on Llama3 architecture and fine-tuned using specialized clinical data. They underwent multi-stage preference alignment to effectively respond to natural prompts. While generic models are often preference-aligned to avoid answering clinical queries as a precaution, Med42-v2 is specifically trained to overcome this limitation, enabling its use in clinical settings. Med42-v2 models demonstrate superior performance compared to the original Llama3 models in both 8B and 70B parameter configurations and GPT-4 across various medical benchmarks. These LLMs are developed to understand clinical queries, perform reasoning tasks, and provide valuable assistance in clinical environments. The models are now publicly available at \href{https://huggingface.co/m42-health}{https://huggingface.co/m42-health}.
Beyond Metrics: A Critical Analysis of the Variability in Large Language Model Evaluation Frameworks
Jul 29, 2024



Abstract:As large language models (LLMs) continue to evolve, the need for robust and standardized evaluation benchmarks becomes paramount. Evaluating the performance of these models is a complex challenge that requires careful consideration of various linguistic tasks, model architectures, and benchmarking methodologies. In recent years, various frameworks have emerged as noteworthy contributions to the field, offering comprehensive evaluation tests and benchmarks for assessing the capabilities of LLMs across diverse domains. This paper provides an exploration and critical analysis of some of these evaluation methodologies, shedding light on their strengths, limitations, and impact on advancing the state-of-the-art in natural language processing.
Med42 -- Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches
Apr 23, 2024



Abstract:This study presents a comprehensive analysis and comparison of two predominant fine-tuning methodologies - full-parameter fine-tuning and parameter-efficient tuning - within the context of medical Large Language Models (LLMs). We developed and refined a series of LLMs, based on the Llama-2 architecture, specifically designed to enhance medical knowledge retrieval, reasoning, and question-answering capabilities. Our experiments systematically evaluate the effectiveness of these tuning strategies across various well-known medical benchmarks. Notably, our medical LLM Med42 showed an accuracy level of 72% on the US Medical Licensing Examination (USMLE) datasets, setting a new standard in performance for openly available medical LLMs. Through this comparative analysis, we aim to identify the most effective and efficient method for fine-tuning LLMs in the medical domain, thereby contributing significantly to the advancement of AI-driven healthcare applications.
Monitoring geometrical properties of word embeddings for detecting the emergence of new topics
Nov 05, 2021



Abstract:Slow emerging topic detection is a task between event detection, where we aggregate behaviors of different words on short period of time, and language evolution, where we monitor their long term evolution. In this work, we tackle the problem of early detection of slowly emerging new topics. To this end, we gather evidence of weak signals at the word level. We propose to monitor the behavior of words representation in an embedding space and use one of its geometrical properties to characterize the emergence of topics. As evaluation is typically hard for this kind of task, we present a framework for quantitative evaluation. We show positive results that outperform state-of-the-art methods on two public datasets of press and scientific articles.
How to detect novelty in textual data streams? A comparative study of existing methods
Sep 11, 2019



Abstract:Since datasets with annotation for novelty at the document and/or word level are not easily available, we present a simulation framework that allows us to create different textual datasets in which we control the way novelty occurs. We also present a benchmark of existing methods for novelty detection in textual data streams. We define a few tasks to solve and compare several state-of-the-art methods. The simulation framework allows us to evaluate their performances according to a set of limited scenarios and test their sensitivity to some parameters. Finally, we experiment with the same methods on different kinds of novelty in the New York Times Annotated Dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge