Shadab Khan
Named Clinical Entity Recognition Benchmark
Oct 07, 2024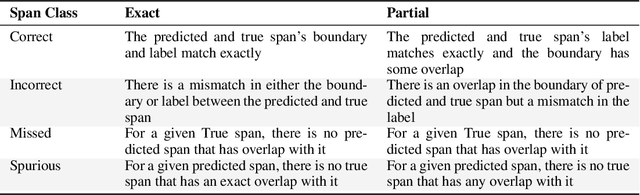
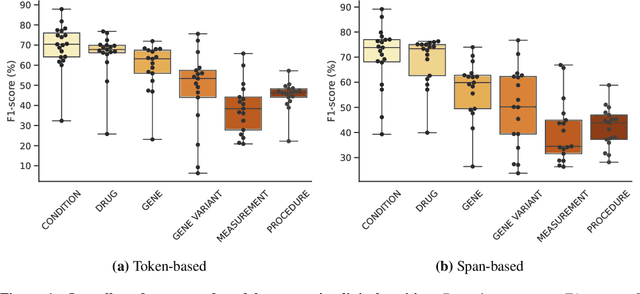
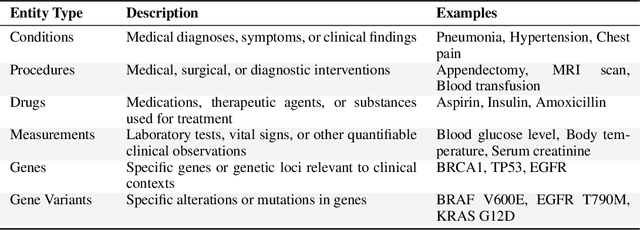
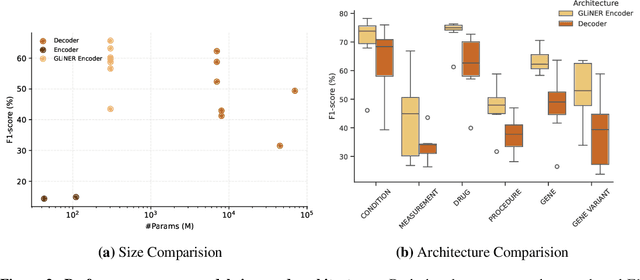
Abstract:This technical report introduces a Named Clinical Entity Recognition Benchmark for evaluating language models in healthcare, addressing the crucial natural language processing (NLP) task of extracting structured information from clinical narratives to support applications like automated coding, clinical trial cohort identification, and clinical decision support. The leaderboard provides a standardized platform for assessing diverse language models, including encoder and decoder architectures, on their ability to identify and classify clinical entities across multiple medical domains. A curated collection of openly available clinical datasets is utilized, encompassing entities such as diseases, symptoms, medications, procedures, and laboratory measurements. Importantly, these entities are standardized according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model, ensuring consistency and interoperability across different healthcare systems and datasets, and a comprehensive evaluation of model performance. Performance of models is primarily assessed using the F1-score, and it is complemented by various assessment modes to provide comprehensive insights into model performance. The report also includes a brief analysis of models evaluated to date, highlighting observed trends and limitations. By establishing this benchmarking framework, the leaderboard aims to promote transparency, facilitate comparative analyses, and drive innovation in clinical entity recognition tasks, addressing the need for robust evaluation methods in healthcare NLP.
MEDIC: Towards a Comprehensive Framework for Evaluating LLMs in Clinical Applications
Sep 11, 2024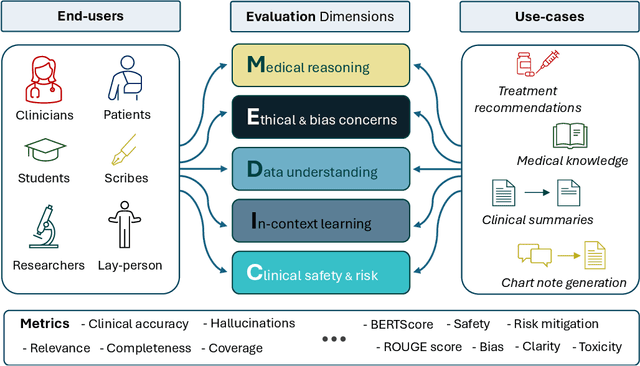
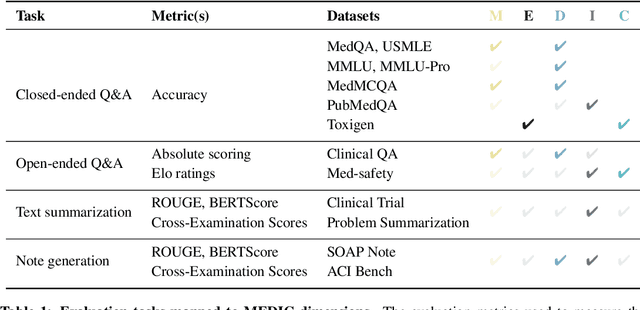
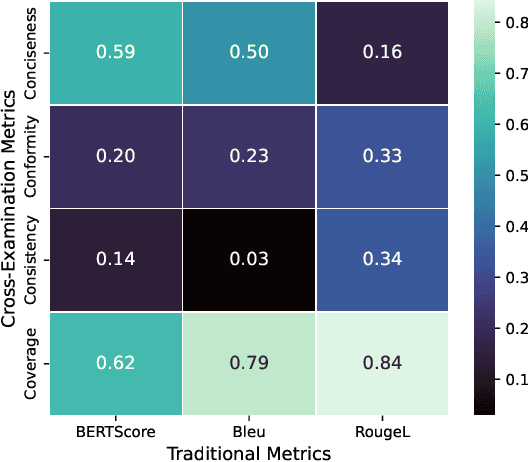
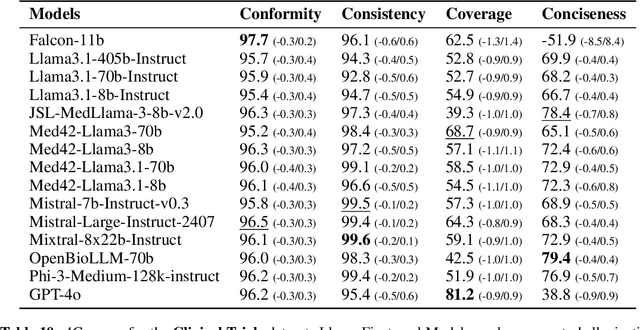
Abstract:The rapid development of Large Language Models (LLMs) for healthcare applications has spurred calls for holistic evaluation beyond frequently-cited benchmarks like USMLE, to better reflect real-world performance. While real-world assessments are valuable indicators of utility, they often lag behind the pace of LLM evolution, likely rendering findings obsolete upon deployment. This temporal disconnect necessitates a comprehensive upfront evaluation that can guide model selection for specific clinical applications. We introduce MEDIC, a framework assessing LLMs across five critical dimensions of clinical competence: medical reasoning, ethics and bias, data and language understanding, in-context learning, and clinical safety. MEDIC features a novel cross-examination framework quantifying LLM performance across areas like coverage and hallucination detection, without requiring reference outputs. We apply MEDIC to evaluate LLMs on medical question-answering, safety, summarization, note generation, and other tasks. Our results show performance disparities across model sizes, baseline vs medically finetuned models, and have implications on model selection for applications requiring specific model strengths, such as low hallucination or lower cost of inference. MEDIC's multifaceted evaluation reveals these performance trade-offs, bridging the gap between theoretical capabilities and practical implementation in healthcare settings, ensuring that the most promising models are identified and adapted for diverse healthcare applications.
Med42-v2: A Suite of Clinical LLMs
Aug 12, 2024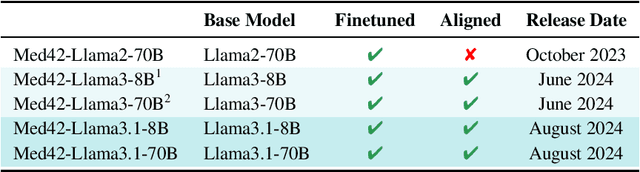
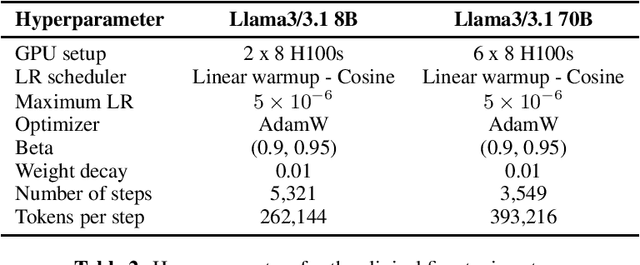
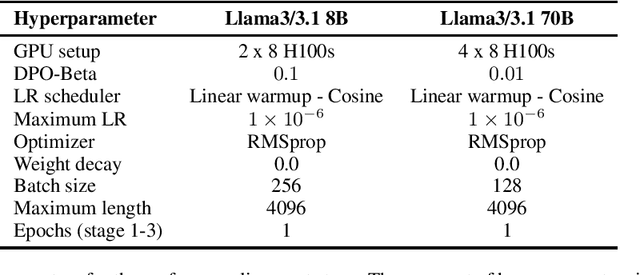
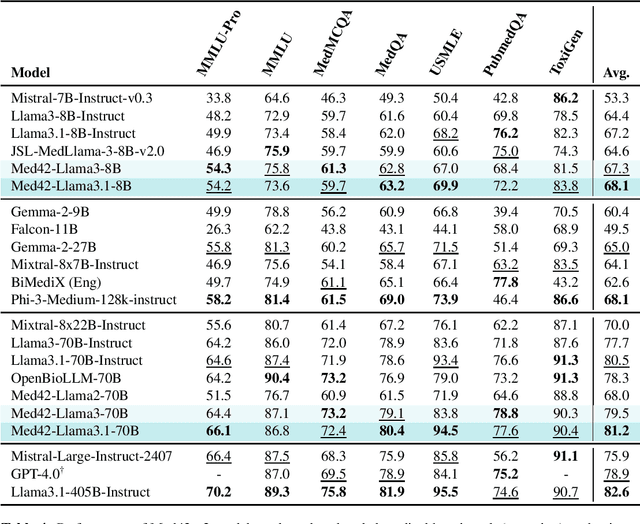
Abstract:Med42-v2 introduces a suite of clinical large language models (LLMs) designed to address the limitations of generic models in healthcare settings. These models are built on Llama3 architecture and fine-tuned using specialized clinical data. They underwent multi-stage preference alignment to effectively respond to natural prompts. While generic models are often preference-aligned to avoid answering clinical queries as a precaution, Med42-v2 is specifically trained to overcome this limitation, enabling its use in clinical settings. Med42-v2 models demonstrate superior performance compared to the original Llama3 models in both 8B and 70B parameter configurations and GPT-4 across various medical benchmarks. These LLMs are developed to understand clinical queries, perform reasoning tasks, and provide valuable assistance in clinical environments. The models are now publicly available at \href{https://huggingface.co/m42-health}{https://huggingface.co/m42-health}.
Beyond Metrics: A Critical Analysis of the Variability in Large Language Model Evaluation Frameworks
Jul 29, 2024



Abstract:As large language models (LLMs) continue to evolve, the need for robust and standardized evaluation benchmarks becomes paramount. Evaluating the performance of these models is a complex challenge that requires careful consideration of various linguistic tasks, model architectures, and benchmarking methodologies. In recent years, various frameworks have emerged as noteworthy contributions to the field, offering comprehensive evaluation tests and benchmarks for assessing the capabilities of LLMs across diverse domains. This paper provides an exploration and critical analysis of some of these evaluation methodologies, shedding light on their strengths, limitations, and impact on advancing the state-of-the-art in natural language processing.
Med42 -- Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches
Apr 23, 2024



Abstract:This study presents a comprehensive analysis and comparison of two predominant fine-tuning methodologies - full-parameter fine-tuning and parameter-efficient tuning - within the context of medical Large Language Models (LLMs). We developed and refined a series of LLMs, based on the Llama-2 architecture, specifically designed to enhance medical knowledge retrieval, reasoning, and question-answering capabilities. Our experiments systematically evaluate the effectiveness of these tuning strategies across various well-known medical benchmarks. Notably, our medical LLM Med42 showed an accuracy level of 72% on the US Medical Licensing Examination (USMLE) datasets, setting a new standard in performance for openly available medical LLMs. Through this comparative analysis, we aim to identify the most effective and efficient method for fine-tuning LLMs in the medical domain, thereby contributing significantly to the advancement of AI-driven healthcare applications.
A machine learning-based method for estimating the number and orientations of major fascicles in diffusion-weighted magnetic resonance imaging
Jun 19, 2020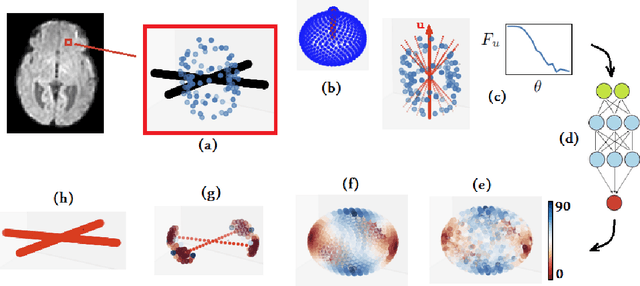
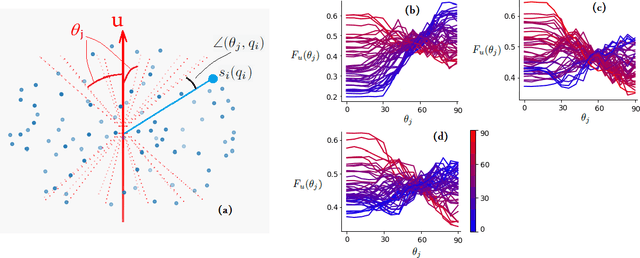
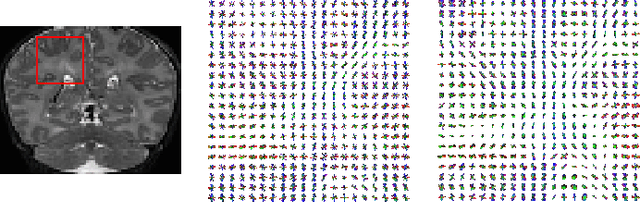
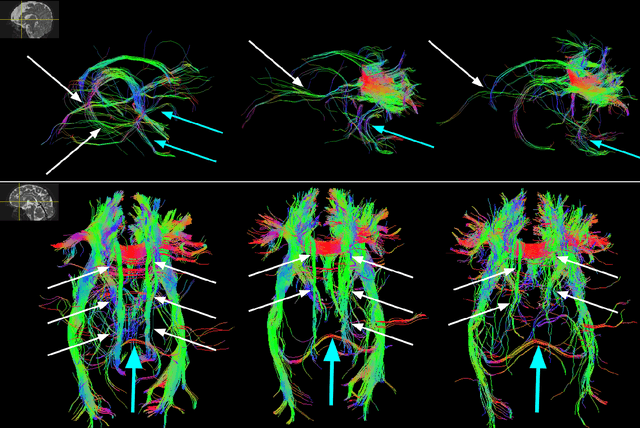
Abstract:Multi-compartment modeling of diffusion-weighted magnetic resonance imaging measurements is necessary for accurate brain connectivity analysis. Existing methods for estimating the number and orientations of fascicles in an imaging voxel either depend on non-convex optimization techniques that are sensitive to initialization and measurement noise, or are prone to predicting spurious fascicles. In this paper, we propose a machine learning-based technique that can accurately estimate the number and orientations of fascicles in a voxel. Our method can be trained with either simulated or real diffusion-weighted imaging data. Our method estimates the angle to the closest fascicle for each direction in a set of discrete directions uniformly spread on the unit sphere. This information is then processed to extract the number and orientations of fascicles in a voxel. On realistic simulated phantom data with known ground truth, our method predicts the number and orientations of crossing fascicles more accurately than several existing methods. It also leads to more accurate tractography. On real data, our method is better than or compares favorably with standard methods in terms of robustness to measurement down-sampling and also in terms of expert quality assessment of tractography results.
FAIRS -- Soft Focus Generator and Attention for Robust Object Segmentation from Extreme Points
Apr 04, 2020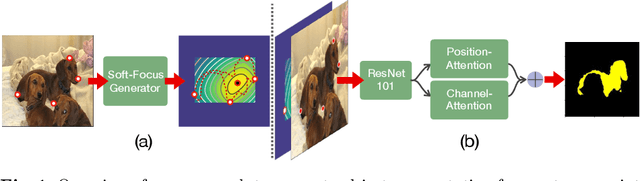
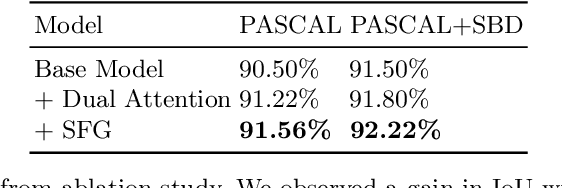
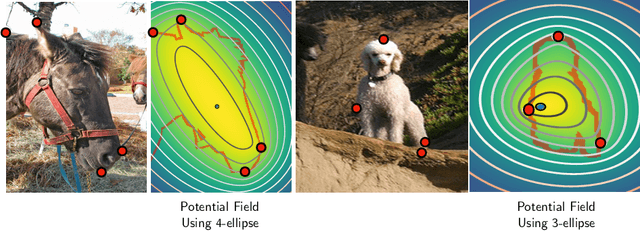
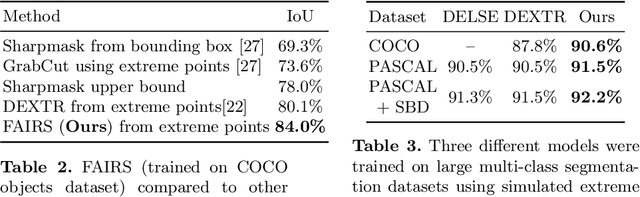
Abstract:Semantic segmentation from user inputs has been actively studied to facilitate interactive segmentation for data annotation and other applications. Recent studies have shown that extreme points can be effectively used to encode user inputs. A heat map generated from the extreme points can be appended to the RGB image and input to the model for training. In this study, we present FAIRS -- a new approach to generate object segmentation from user inputs in the form of extreme points and corrective clicks. We propose a novel approach for effectively encoding the user input from extreme points and corrective clicks, in a novel and scalable manner that allows the network to work with a variable number of clicks, including corrective clicks for output refinement. We also integrate a dual attention module with our approach to increase the efficacy of the model in preferentially attending to the objects. We demonstrate that these additions help achieve significant improvements over state-of-the-art in dense object segmentation from user inputs, on multiple large-scale datasets. Through experiments, we demonstrate our method's ability to generate high-quality training data as well as its scalability in incorporating extreme points, guiding clicks, and corrective clicks in a principled manner.
Towards Robust and Reproducible Active Learning Using Neural Networks
Feb 21, 2020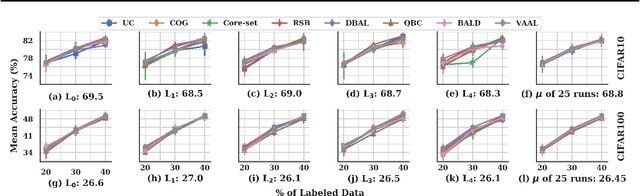
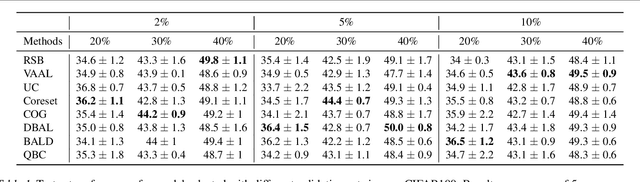
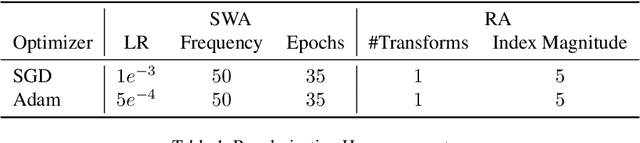
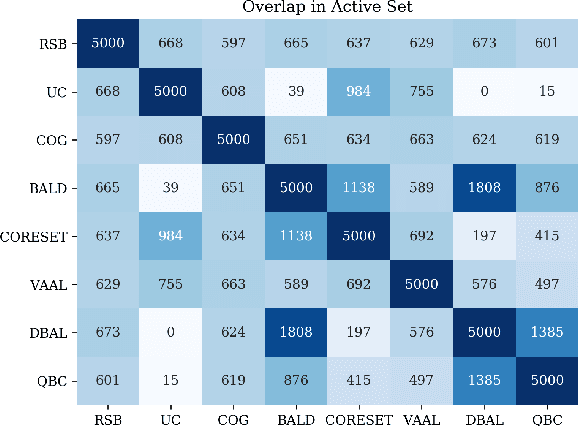
Abstract:Active learning (AL) is a promising ML paradigm that has the potential to parse through large unlabeled data and help reduce annotation cost in domains where labeling entire data can be prohibitive. Recently proposed neural network based AL methods use different heuristics to accomplish this goal. In this study, we show that recent AL methods offer a gain over random baseline under a brittle combination of experimental conditions. We demonstrate that such marginal gains vanish when experimental factors are changed, leading to reproducibility issues and suggesting that AL methods lack robustness. We also observe that with a properly tuned model, which employs recently proposed regularization techniques, the performance significantly improves for all AL methods including the random sampling baseline, and performance differences among the AL methods become negligible. Based on these observations, we suggest a set of experiments that are critical to assess the true effectiveness of an AL method. To facilitate these experiments we also present an open source toolkit. We believe our findings and recommendations will help advance reproducible research in robust AL using neural networks.
Extreme Points Derived Confidence Map as a Cue For Class-Agnostic Segmentation Using Deep Neural Network
Jun 06, 2019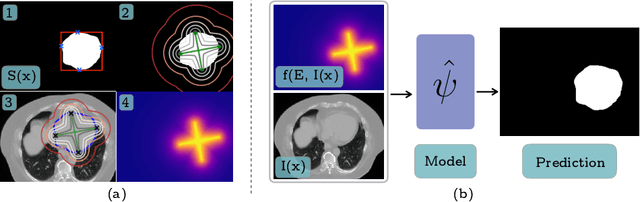
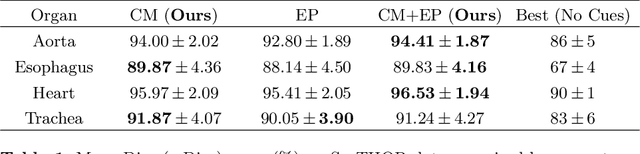
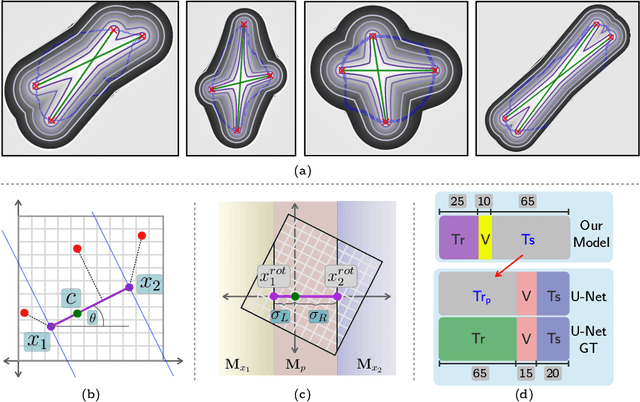
Abstract:To automate the process of segmenting an anatomy of interest, we can learn a model from previously annotated data. The learning-based approach uses annotations to train a model that tries to emulate the expert labeling on a new data set. While tremendous progress has been made using such approaches, labeling of medical images remains a time-consuming and expensive task. In this paper, we evaluate the utility of extreme points in learning to segment. Specifically, we propose a novel approach to compute a confidence map from extreme points that quantitatively encodes the priors derived from extreme points. We use the confidence map as a cue to train a deep neural network based on ResNet-101 and PSP module to develop a class-agnostic segmentation model that outperforms state-of-the-art method that employs extreme points as a cue. Further, we evaluate a realistic use-case by using our model to generate training data for supervised learning (U-Net) and observed that U-Net performs comparably when trained with either the generated data or the ground truth data. These findings suggest that models trained using cues can be used to generate reliable training data.
Real-time Deep Pose Estimation with Geodesic Loss for Image-to-Template Rigid Registration
Aug 18, 2018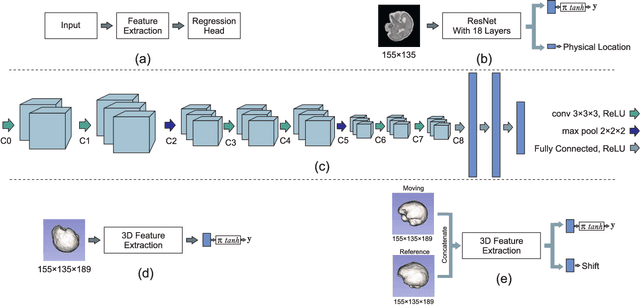
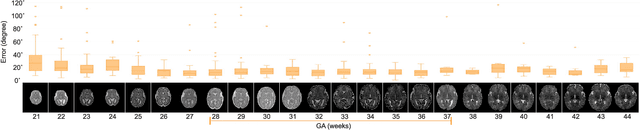
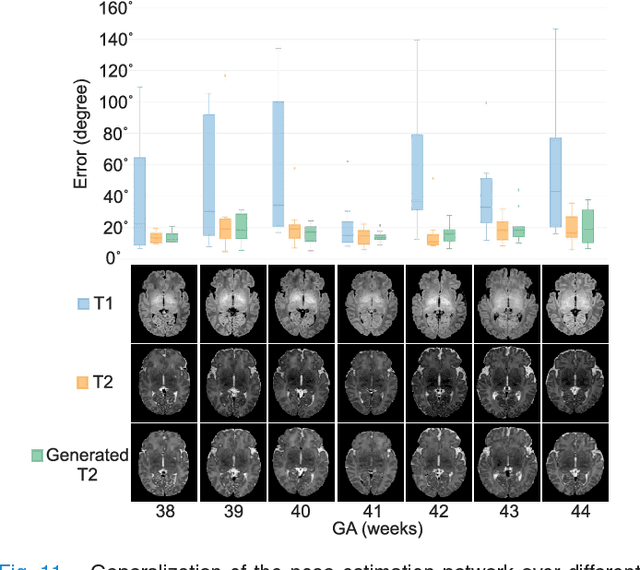
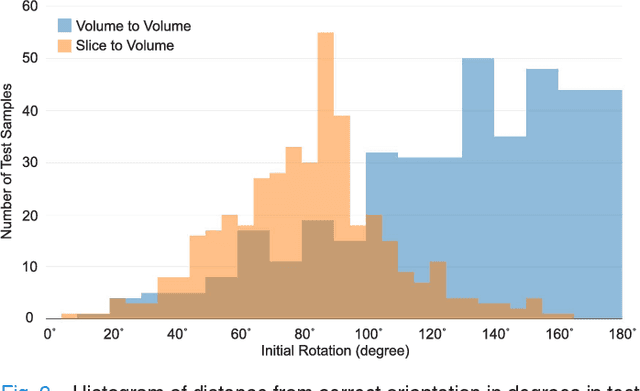
Abstract:With an aim to increase the capture range and accelerate the performance of state-of-the-art inter-subject and subject-to-template 3D registration, we propose deep learning-based methods that are trained to find the 3D position of arbitrarily oriented subjects or anatomy based on slices or volumes of medical images. For this, we propose regression CNNs that learn to predict the angle-axis representation of 3D rotations and translations using image features. We use and compare mean square error and geodesic loss to train regression CNNs for 3D pose estimation used in two different scenarios: slice-to-volume registration and volume-to-volume registration. Our results show that in such registration applications that are amendable to learning, the proposed deep learning methods with geodesic loss minimization can achieve accurate results with a wide capture range in real-time (<100ms). We also tested the generalization capability of the trained CNNs on an expanded age range and on images of newborn subjects with similar and different MR image contrasts. We trained our models on T2-weighted fetal brain MRI scans and used them to predict the 3D pose of newborn brains based on T1-weighted MRI scans. We showed that the trained models generalized well for the new domain when we performed image contrast transfer through a conditional generative adversarial network. This indicates that the domain of application of the trained deep regression CNNs can be further expanded to image modalities and contrasts other than those used in training. A combination of our proposed methods with accelerated optimization-based registration algorithms can dramatically enhance the performance of automatic imaging devices and image processing methods of the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge