Lana Vasung
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024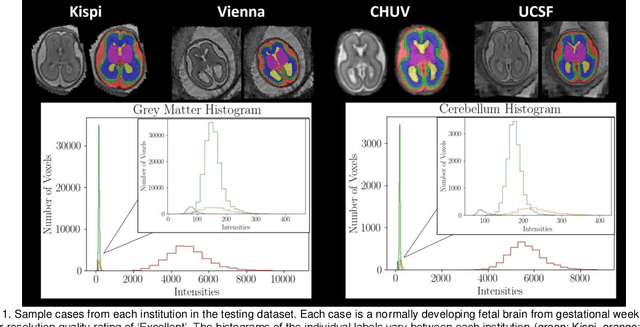
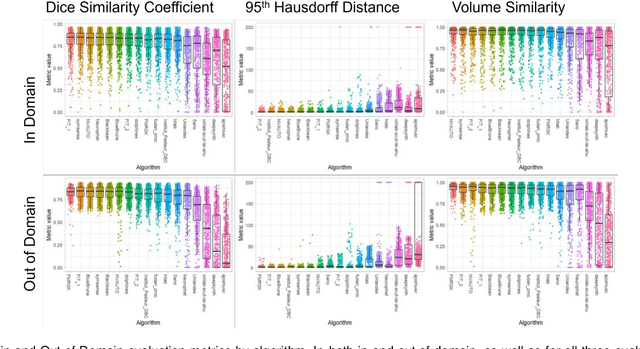
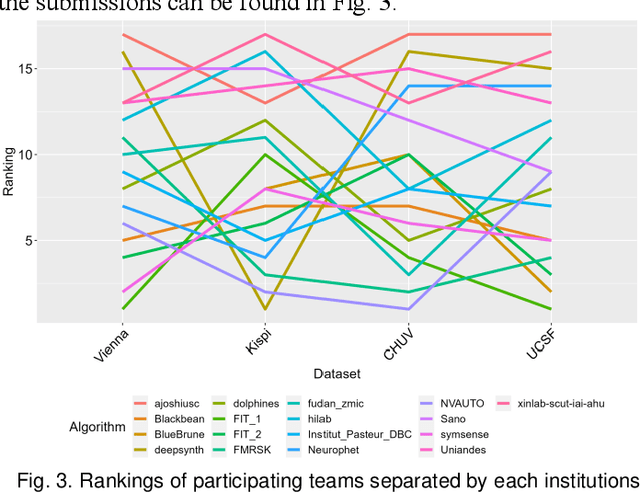
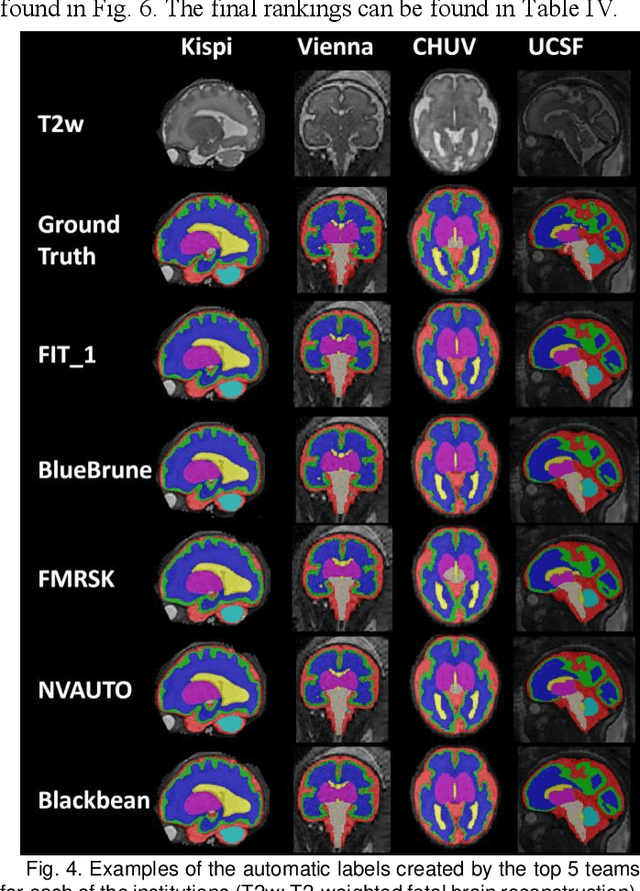
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Characterizing normal perinatal development of the human brain structural connectivity
Aug 22, 2023Abstract:Early brain development is characterized by the formation of a highly organized structural connectome. The interconnected nature of this connectome underlies the brain's cognitive abilities and influences its response to diseases and environmental factors. Hence, quantitative assessment of structural connectivity in the perinatal stage is useful for studying normal and abnormal neurodevelopment. However, estimation of the connectome from diffusion MRI data involves complex computations. For the perinatal period, these computations are further challenged by the rapid brain development and imaging difficulties. Combined with high inter-subject variability, these factors make it difficult to chart the normal development of the structural connectome. As a result, there is a lack of reliable normative baselines of structural connectivity metrics at this critical stage in brain development. In this study, we developed a computational framework, based on spatio-temporal averaging, for determining such baselines. We used this framework to analyze the structural connectivity between 33 and 44 postmenstrual weeks using data from 166 subjects. Our results unveiled clear and strong trends in the development of structural connectivity in perinatal stage. Connection weighting based on fractional anisotropy and neurite density produced the most consistent results. We observed increases in global and local efficiency, a decrease in characteristic path length, and widespread strengthening of the connections within and across brain lobes and hemispheres. We also observed asymmetry patterns that were consistent between different connection weighting approaches. The new computational method and results are useful for assessing normal and abnormal development of the structural connectome early in life.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022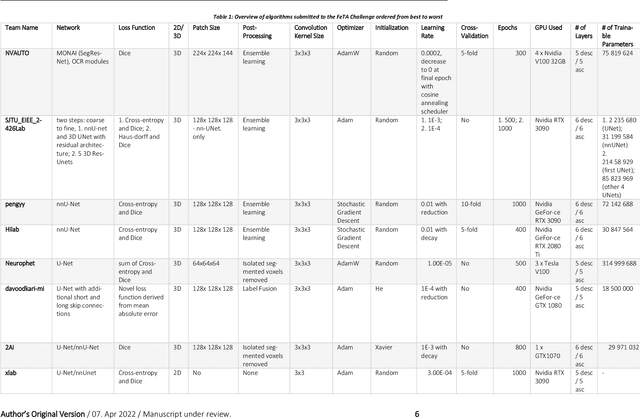
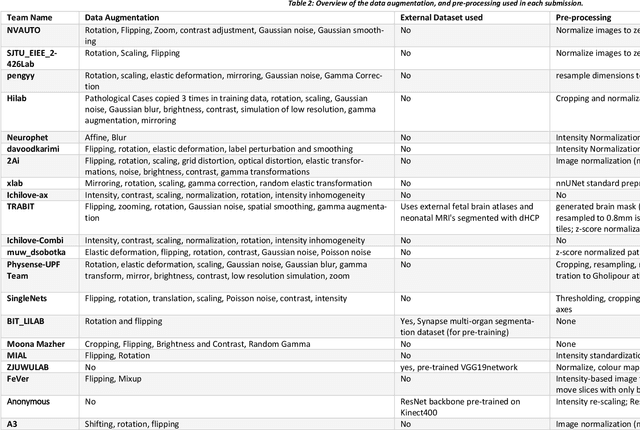
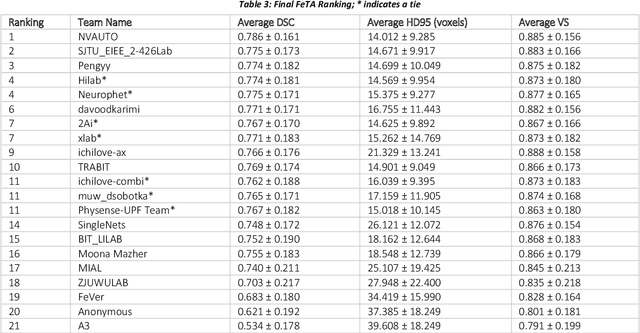
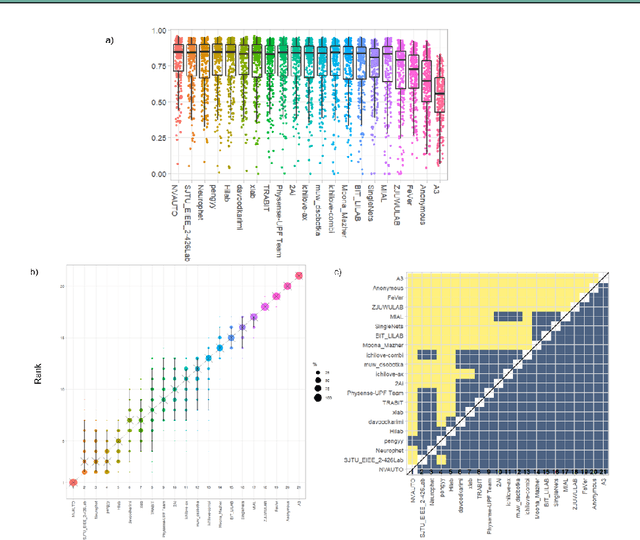
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
A machine learning-based method for estimating the number and orientations of major fascicles in diffusion-weighted magnetic resonance imaging
Jun 19, 2020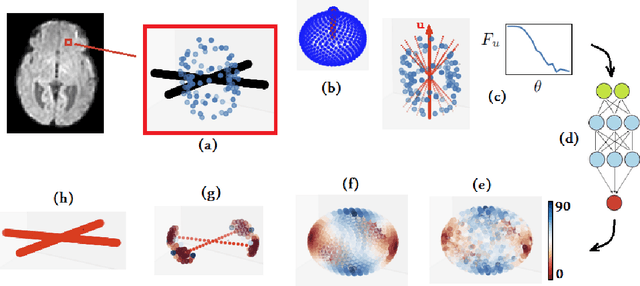
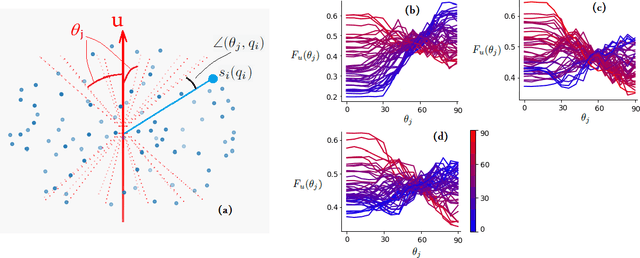
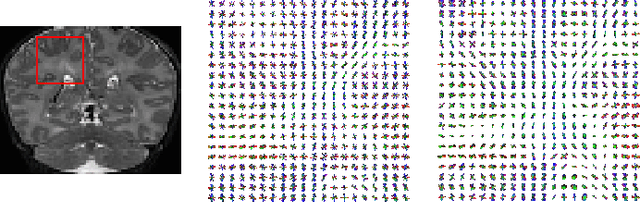
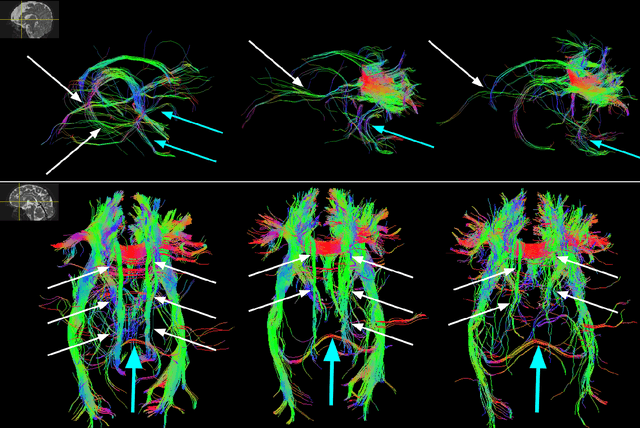
Abstract:Multi-compartment modeling of diffusion-weighted magnetic resonance imaging measurements is necessary for accurate brain connectivity analysis. Existing methods for estimating the number and orientations of fascicles in an imaging voxel either depend on non-convex optimization techniques that are sensitive to initialization and measurement noise, or are prone to predicting spurious fascicles. In this paper, we propose a machine learning-based technique that can accurately estimate the number and orientations of fascicles in a voxel. Our method can be trained with either simulated or real diffusion-weighted imaging data. Our method estimates the angle to the closest fascicle for each direction in a set of discrete directions uniformly spread on the unit sphere. This information is then processed to extract the number and orientations of fascicles in a voxel. On realistic simulated phantom data with known ground truth, our method predicts the number and orientations of crossing fascicles more accurately than several existing methods. It also leads to more accurate tractography. On real data, our method is better than or compares favorably with standard methods in terms of robustness to measurement down-sampling and also in terms of expert quality assessment of tractography results.
A Deep Attentive Convolutional Neural Network for Automatic Cortical Plate Segmentation in Fetal MRI
Apr 27, 2020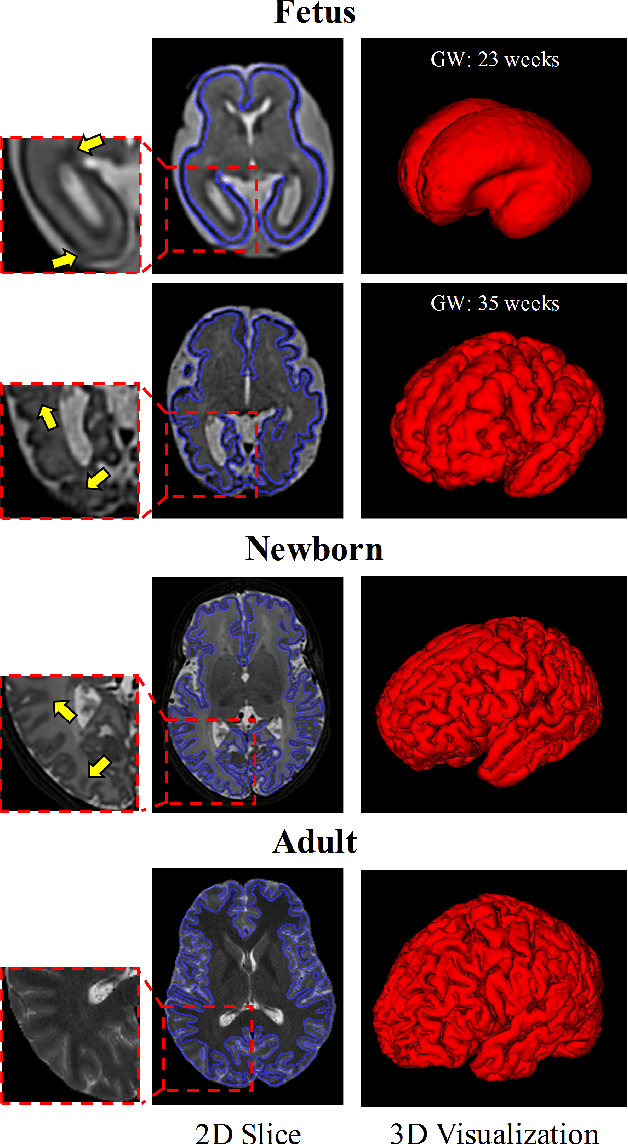
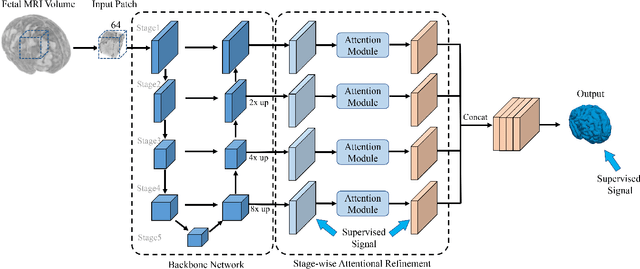
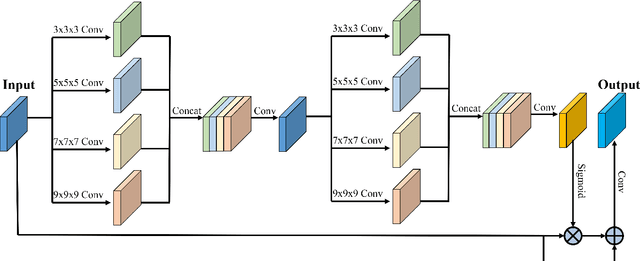
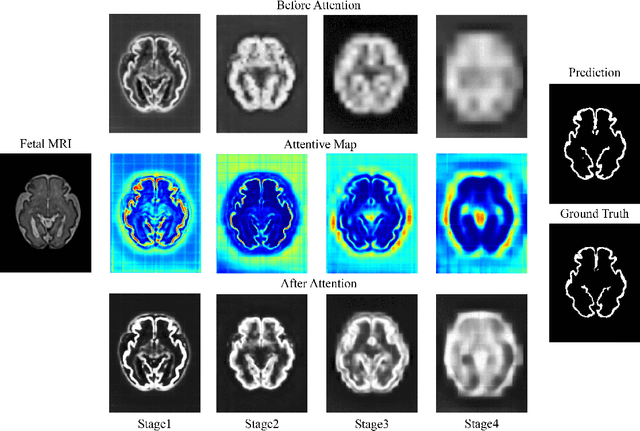
Abstract:Fetal cortical plate segmentation is essential in quantitative analysis of fetal brain maturation and cortical folding. Manual segmentation of the cortical plate, or manual refinement of automatic segmentations is tedious and time consuming, and automatic segmentation of the cortical plate is challenged by the relatively low resolution of the reconstructed fetal brain MRI scans compared to the thin structure of the cortical plate, partial voluming, and the wide range of variations in the morphology of the cortical plate as the brain matures during gestation. To reduce the burden of manual refinement of segmentations, we have developed a new and powerful deep learning segmentation method that exploits new deep attentive modules with mixed kernel convolutions within a fully convolutional neural network architecture that utilizes deep supervision and residual connections. Quantitative evaluation based on several performance measures and expert evaluations show that our method outperformed several state-of-the-art deep models for segmentation, as well as a state-of-the-art multi-atlas segmentation technique. In particular, we achieved average Dice similarity coefficient of 0.87, average Hausdroff distance of 0.96mm, and average symmetric surface difference of 0.28mm in cortical plate segmentation on reconstructed fetal brain MRI scans of fetuses scanned in the gestational age range of 16 to 39 weeks. By generating accurate cortical plate segmentations in less than 2 minutes, our method can facilitate and accelerate large-scale studies on normal and altered fetal brain cortical maturation and folding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge