Caitlin K. Rollins
Learning to segment fetal brain tissue from noisy annotations
Mar 25, 2022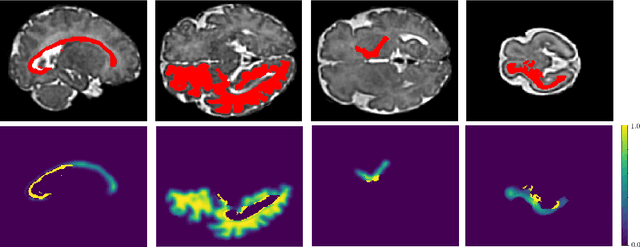
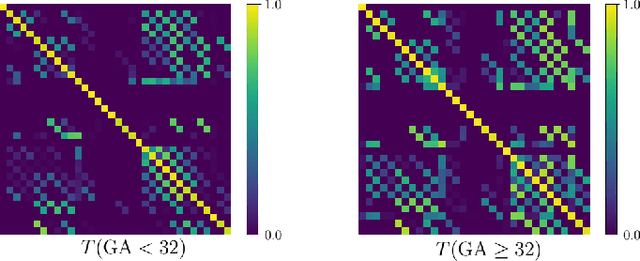
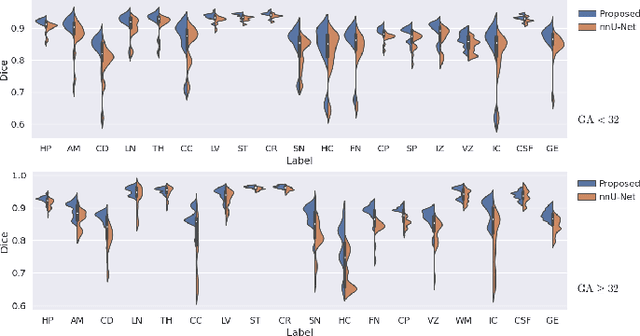
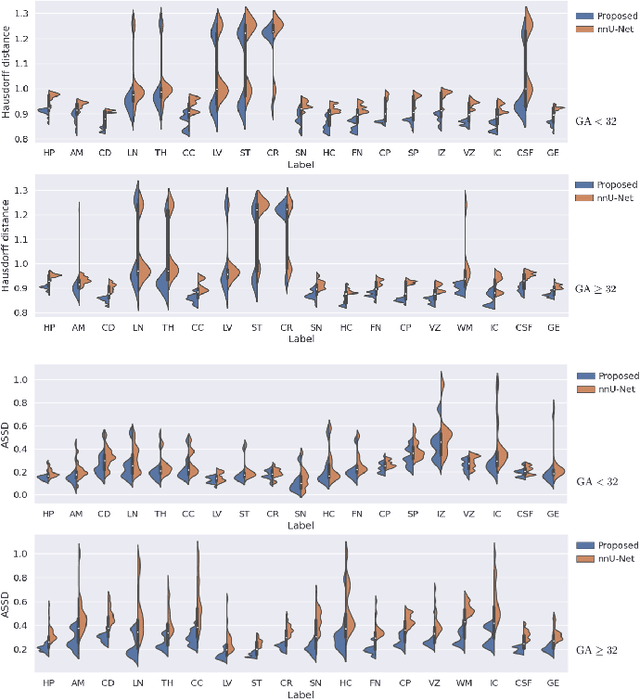
Abstract:Automatic fetal brain tissue segmentation can enhance the quantitative assessment of brain development at this critical stage. Deep learning methods represent the state of the art in medical image segmentation and have also achieved impressive results in brain segmentation. However, effective training of a deep learning model to perform this task requires a large number of training images to represent the rapid development of the transient fetal brain structures. On the other hand, manual multi-label segmentation of a large number of 3D images is prohibitive. To address this challenge, we segmented 272 training images, covering 19-39 gestational weeks, using an automatic multi-atlas segmentation strategy based on deformable registration and probabilistic atlas fusion, and manually corrected large errors in those segmentations. Since this process generated a large training dataset with noisy segmentations, we developed a novel label smoothing procedure and a loss function to train a deep learning model with smoothed noisy segmentations. Our proposed methods properly account for the uncertainty in tissue boundaries. We evaluated our method on 23 manually-segmented test images of a separate set of fetuses. Results show that our method achieves an average Dice similarity coefficient of 0.893 and 0.916 for the transient structures of younger and older fetuses, respectively. Our method generated results that were significantly more accurate than several state-of-the-art methods including nnU-Net that achieved the closest results to our method. Our trained model can serve as a valuable tool to enhance the accuracy and reproducibility of fetal brain analysis in MRI.
A Deep Attentive Convolutional Neural Network for Automatic Cortical Plate Segmentation in Fetal MRI
Apr 27, 2020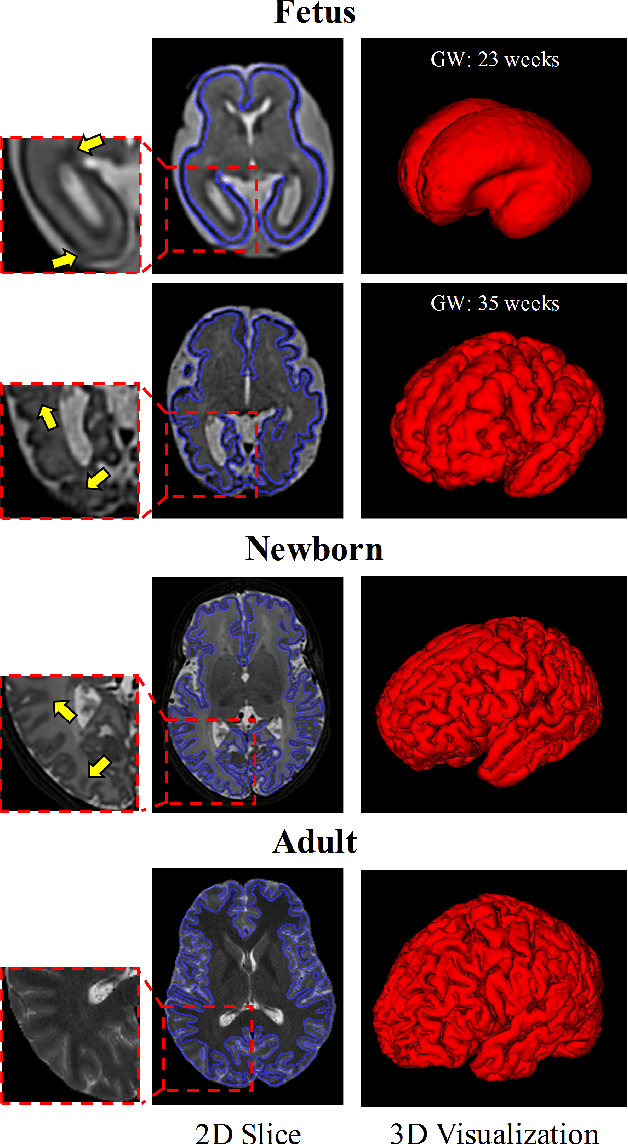
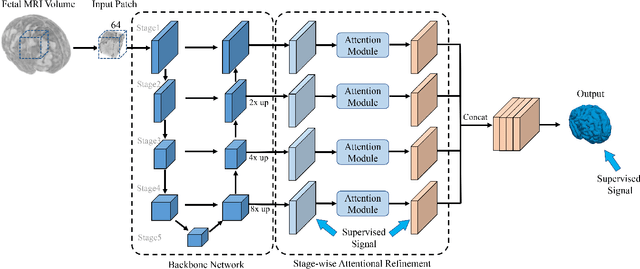
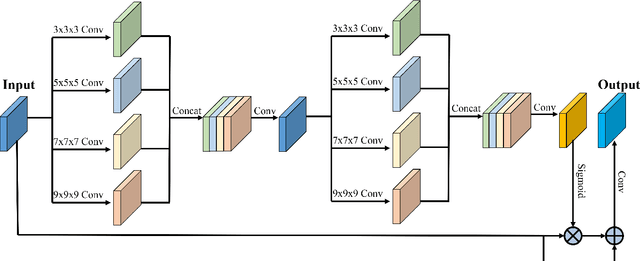
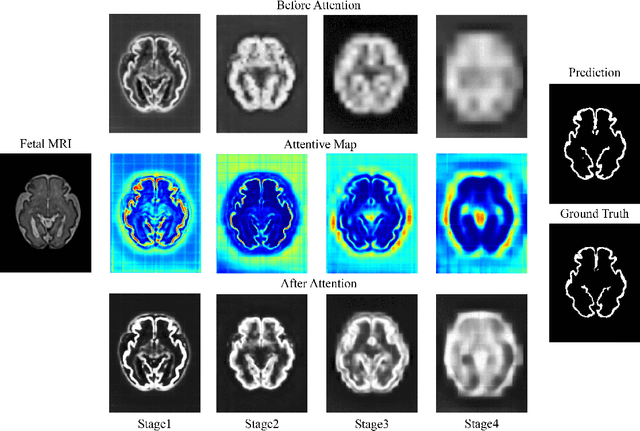
Abstract:Fetal cortical plate segmentation is essential in quantitative analysis of fetal brain maturation and cortical folding. Manual segmentation of the cortical plate, or manual refinement of automatic segmentations is tedious and time consuming, and automatic segmentation of the cortical plate is challenged by the relatively low resolution of the reconstructed fetal brain MRI scans compared to the thin structure of the cortical plate, partial voluming, and the wide range of variations in the morphology of the cortical plate as the brain matures during gestation. To reduce the burden of manual refinement of segmentations, we have developed a new and powerful deep learning segmentation method that exploits new deep attentive modules with mixed kernel convolutions within a fully convolutional neural network architecture that utilizes deep supervision and residual connections. Quantitative evaluation based on several performance measures and expert evaluations show that our method outperformed several state-of-the-art deep models for segmentation, as well as a state-of-the-art multi-atlas segmentation technique. In particular, we achieved average Dice similarity coefficient of 0.87, average Hausdroff distance of 0.96mm, and average symmetric surface difference of 0.28mm in cortical plate segmentation on reconstructed fetal brain MRI scans of fetuses scanned in the gestational age range of 16 to 39 weeks. By generating accurate cortical plate segmentations in less than 2 minutes, our method can facilitate and accelerate large-scale studies on normal and altered fetal brain cortical maturation and folding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge