Nasir Hayat
Named Clinical Entity Recognition Benchmark
Oct 07, 2024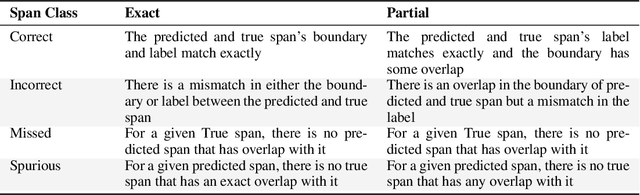
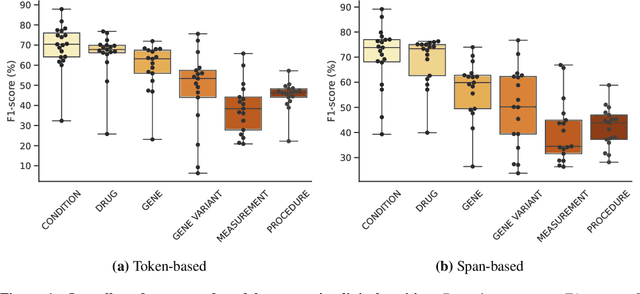
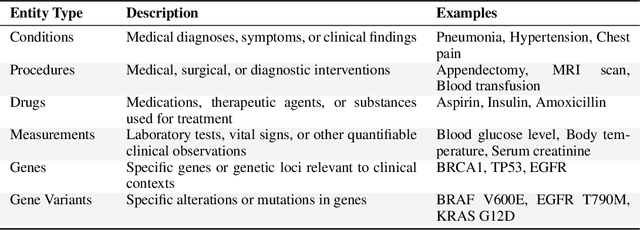
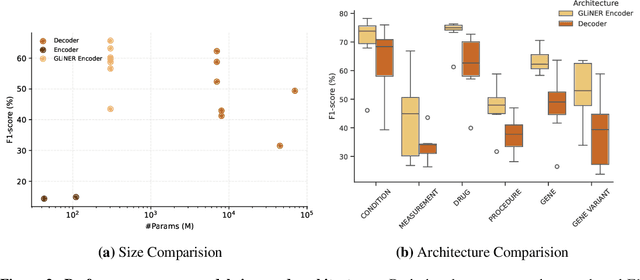
Abstract:This technical report introduces a Named Clinical Entity Recognition Benchmark for evaluating language models in healthcare, addressing the crucial natural language processing (NLP) task of extracting structured information from clinical narratives to support applications like automated coding, clinical trial cohort identification, and clinical decision support. The leaderboard provides a standardized platform for assessing diverse language models, including encoder and decoder architectures, on their ability to identify and classify clinical entities across multiple medical domains. A curated collection of openly available clinical datasets is utilized, encompassing entities such as diseases, symptoms, medications, procedures, and laboratory measurements. Importantly, these entities are standardized according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model, ensuring consistency and interoperability across different healthcare systems and datasets, and a comprehensive evaluation of model performance. Performance of models is primarily assessed using the F1-score, and it is complemented by various assessment modes to provide comprehensive insights into model performance. The report also includes a brief analysis of models evaluated to date, highlighting observed trends and limitations. By establishing this benchmarking framework, the leaderboard aims to promote transparency, facilitate comparative analyses, and drive innovation in clinical entity recognition tasks, addressing the need for robust evaluation methods in healthcare NLP.
MEDIC: Towards a Comprehensive Framework for Evaluating LLMs in Clinical Applications
Sep 11, 2024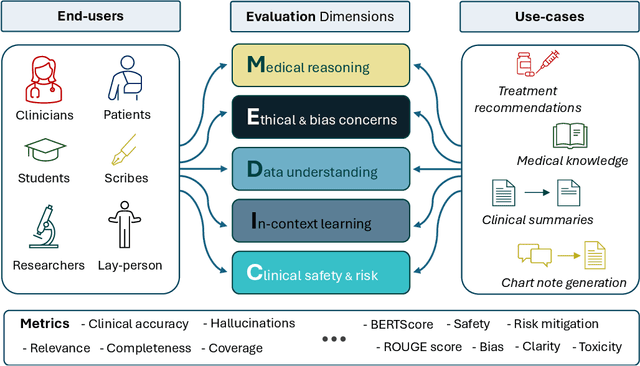
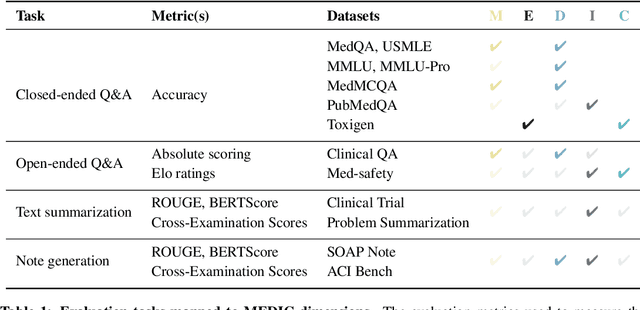
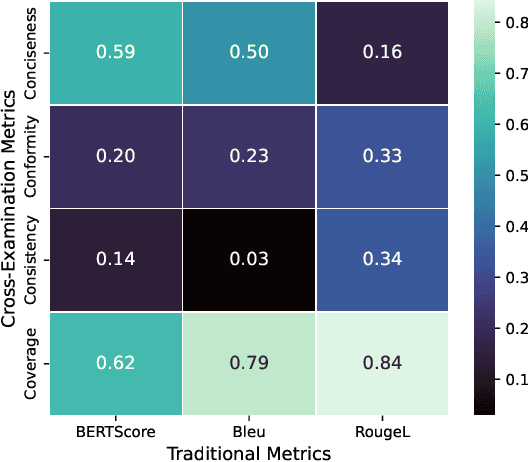
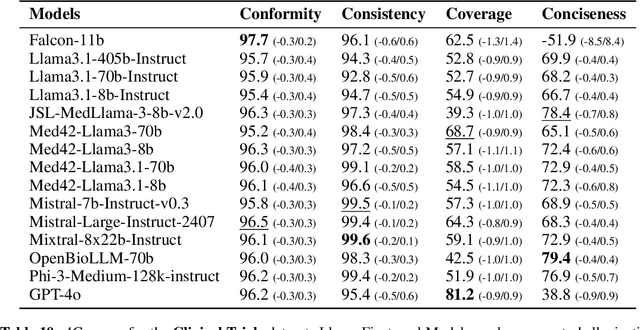
Abstract:The rapid development of Large Language Models (LLMs) for healthcare applications has spurred calls for holistic evaluation beyond frequently-cited benchmarks like USMLE, to better reflect real-world performance. While real-world assessments are valuable indicators of utility, they often lag behind the pace of LLM evolution, likely rendering findings obsolete upon deployment. This temporal disconnect necessitates a comprehensive upfront evaluation that can guide model selection for specific clinical applications. We introduce MEDIC, a framework assessing LLMs across five critical dimensions of clinical competence: medical reasoning, ethics and bias, data and language understanding, in-context learning, and clinical safety. MEDIC features a novel cross-examination framework quantifying LLM performance across areas like coverage and hallucination detection, without requiring reference outputs. We apply MEDIC to evaluate LLMs on medical question-answering, safety, summarization, note generation, and other tasks. Our results show performance disparities across model sizes, baseline vs medically finetuned models, and have implications on model selection for applications requiring specific model strengths, such as low hallucination or lower cost of inference. MEDIC's multifaceted evaluation reveals these performance trade-offs, bridging the gap between theoretical capabilities and practical implementation in healthcare settings, ensuring that the most promising models are identified and adapted for diverse healthcare applications.
Med42 -- Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches
Apr 23, 2024



Abstract:This study presents a comprehensive analysis and comparison of two predominant fine-tuning methodologies - full-parameter fine-tuning and parameter-efficient tuning - within the context of medical Large Language Models (LLMs). We developed and refined a series of LLMs, based on the Llama-2 architecture, specifically designed to enhance medical knowledge retrieval, reasoning, and question-answering capabilities. Our experiments systematically evaluate the effectiveness of these tuning strategies across various well-known medical benchmarks. Notably, our medical LLM Med42 showed an accuracy level of 72% on the US Medical Licensing Examination (USMLE) datasets, setting a new standard in performance for openly available medical LLMs. Through this comparative analysis, we aim to identify the most effective and efficient method for fine-tuning LLMs in the medical domain, thereby contributing significantly to the advancement of AI-driven healthcare applications.
MedFuse: Multi-modal fusion with clinical time-series data and chest X-ray images
Jul 14, 2022
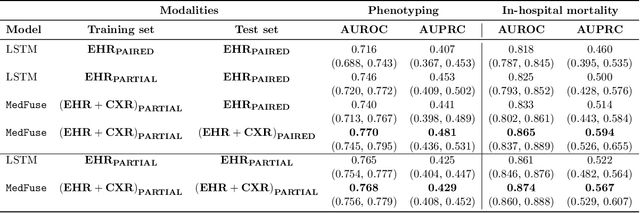

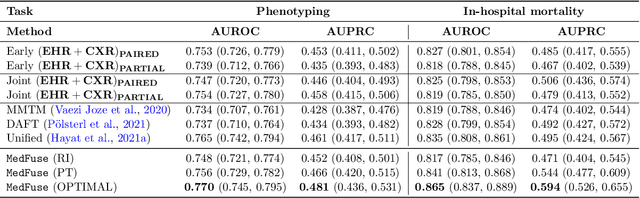
Abstract:Multi-modal fusion approaches aim to integrate information from different data sources. Unlike natural datasets, such as in audio-visual applications, where samples consist of "paired" modalities, data in healthcare is often collected asynchronously. Hence, requiring the presence of all modalities for a given sample is not realistic for clinical tasks and significantly limits the size of the dataset during training. In this paper, we propose MedFuse, a conceptually simple yet promising LSTM-based fusion module that can accommodate uni-modal as well as multi-modal input. We evaluate the fusion method and introduce new benchmark results for in-hospital mortality prediction and phenotype classification, using clinical time-series data in the MIMIC-IV dataset and corresponding chest X-ray images in MIMIC-CXR. Compared to more complex multi-modal fusion strategies, MedFuse provides a performance improvement by a large margin on the fully paired test set. It also remains robust across the partially paired test set containing samples with missing chest X-ray images. We release our code for reproducibility and to enable the evaluation of competing models in the future.
Towards dynamic multi-modal phenotyping using chest radiographs and physiological data
Nov 04, 2021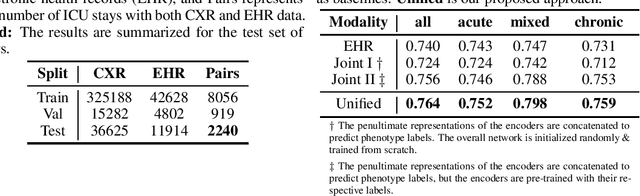
Abstract:The healthcare domain is characterized by heterogeneous data modalities, such as imaging and physiological data. In practice, the variety of medical data assists clinicians in decision-making. However, most of the current state-of-the-art deep learning models solely rely upon carefully curated data of a single modality. In this paper, we propose a dynamic training approach to learn modality-specific data representations and to integrate auxiliary features, instead of solely relying on a single modality. Our preliminary experiments results for a patient phenotyping task using physiological data in MIMIC-IV & chest radiographs in the MIMIC- CXR dataset show that our proposed approach achieves the highest area under the receiver operating characteristic curve (AUROC) (0.764 AUROC) compared to the performance of the benchmark method in previous work, which only used physiological data (0.740 AUROC). For a set of five recurring or chronic diseases with periodic acute episodes, including cardiac dysrhythmia, conduction disorders, and congestive heart failure, the AUROC improves from 0.747 to 0.798. This illustrates the benefit of leveraging the chest imaging modality in the phenotyping task and highlights the potential of multi-modal learning in medical applications.
Multi-Label Generalized Zero Shot Learning for the Classification of Disease in Chest Radiographs
Jul 14, 2021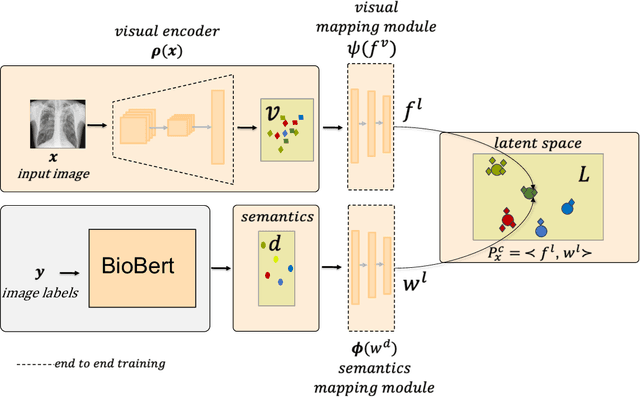
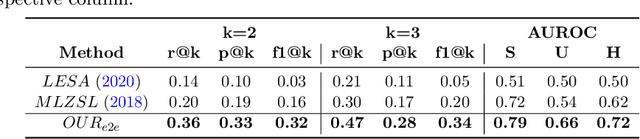
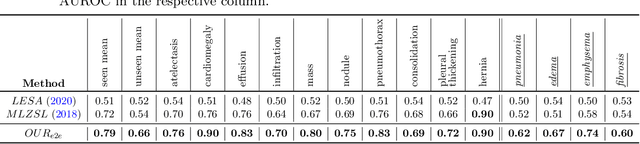
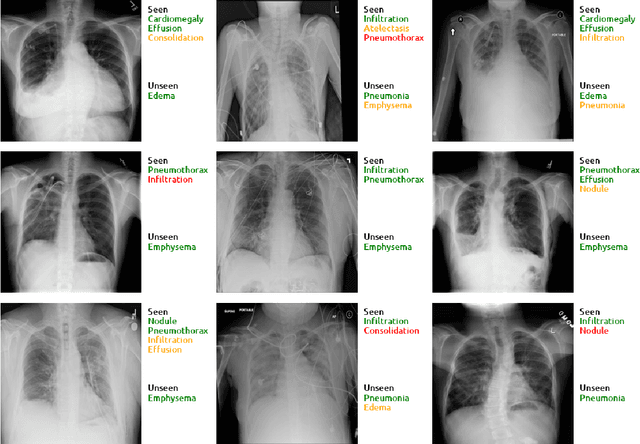
Abstract:Despite the success of deep neural networks in chest X-ray (CXR) diagnosis, supervised learning only allows the prediction of disease classes that were seen during training. At inference, these networks cannot predict an unseen disease class. Incorporating a new class requires the collection of labeled data, which is not a trivial task, especially for less frequently-occurring diseases. As a result, it becomes inconceivable to build a model that can diagnose all possible disease classes. Here, we propose a multi-label generalized zero shot learning (CXR-ML-GZSL) network that can simultaneously predict multiple seen and unseen diseases in CXR images. Given an input image, CXR-ML-GZSL learns a visual representation guided by the input's corresponding semantics extracted from a rich medical text corpus. Towards this ambitious goal, we propose to map both visual and semantic modalities to a latent feature space using a novel learning objective. The objective ensures that (i) the most relevant labels for the query image are ranked higher than irrelevant labels, (ii) the network learns a visual representation that is aligned with its semantics in the latent feature space, and (iii) the mapped semantics preserve their original inter-class representation. The network is end-to-end trainable and requires no independent pre-training for the offline feature extractor. Experiments on the NIH Chest X-ray dataset show that our network outperforms two strong baselines in terms of recall, precision, f1 score, and area under the receiver operating characteristic curve. Our code is publicly available at: https://github.com/nyuad-cai/CXR-ML-GZSL.git
Clinical prediction system of complications among COVID-19 patients: a development and validation retrospective multicentre study
Nov 28, 2020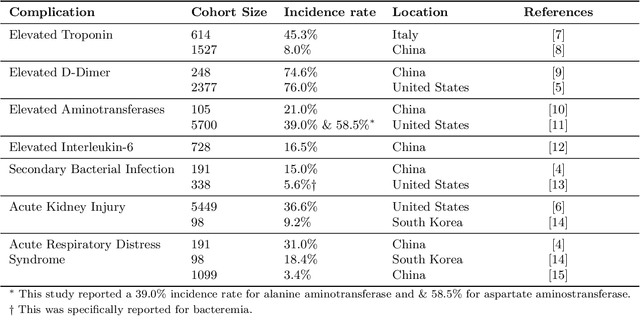


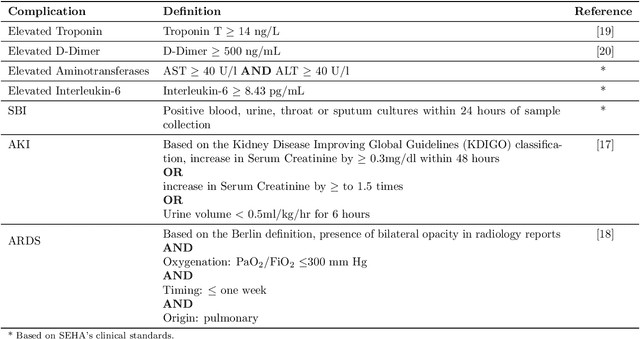
Abstract:Existing prognostic tools mainly focus on predicting the risk of mortality among patients with coronavirus disease 2019. However, clinical evidence suggests that COVID-19 can result in non-mortal complications that affect patient prognosis. To support patient risk stratification, we aimed to develop a prognostic system that predicts complications common to COVID-19. In this retrospective study, we used data collected from 3,352 COVID-19 patient encounters admitted to 18 facilities between April 1 and April 30, 2020, in Abu Dhabi (AD), UAE. The hospitals were split based on geographical proximity to assess for our proposed system's learning generalizability, AD Middle region and AD Western & Eastern regions, A and B, respectively. Using data collected during the first 24 hours of admission, the machine learning-based prognostic system predicts the risk of developing any of seven complications during the hospital stay. The complications include secondary bacterial infection, AKI, ARDS, and elevated biomarkers linked to increased patient severity, including d-dimer, interleukin-6, aminotransferases, and troponin. During training, the system applies an exclusion criteria, hyperparameter tuning, and model selection for each complication-specific model. The system achieves good accuracy across all complications and both regions. In test set A (587 patient encounters), the system achieves 0.91 AUROC for AKI and >0.80 AUROC for most of the other complications. In test set B (225 patient encounters), the respective system achieves 0.90 AUROC for AKI, elevated troponin, and elevated interleukin-6, and >0.80 AUROC for most of the other complications. The best performing models, as selected by our system, were mainly gradient boosting models and logistic regression. Our results show that a data-driven approach using machine learning can predict the risk of such complications with high accuracy.
Synthesizing the Unseen for Zero-shot Object Detection
Oct 19, 2020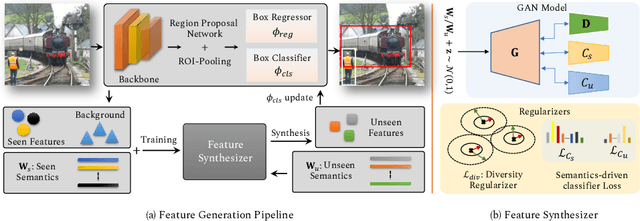
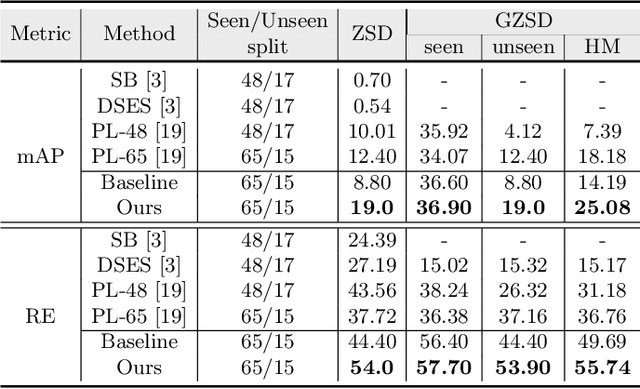
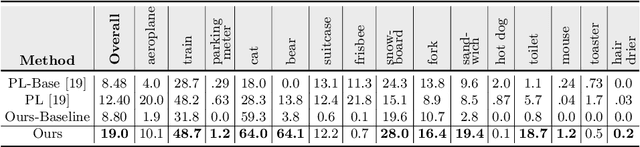
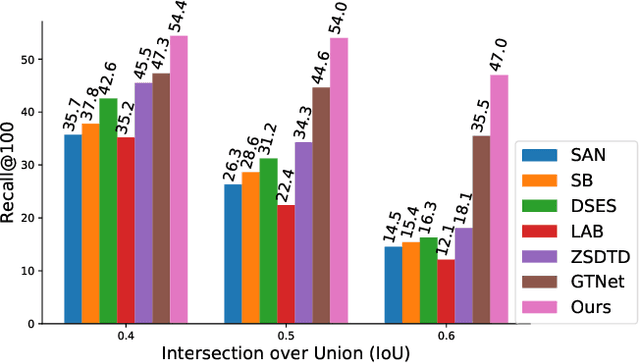
Abstract:The existing zero-shot detection approaches project visual features to the semantic domain for seen objects, hoping to map unseen objects to their corresponding semantics during inference. However, since the unseen objects are never visualized during training, the detection model is skewed towards seen content, thereby labeling unseen as background or a seen class. In this work, we propose to synthesize visual features for unseen classes, so that the model learns both seen and unseen objects in the visual domain. Consequently, the major challenge becomes, how to accurately synthesize unseen objects merely using their class semantics? Towards this ambitious goal, we propose a novel generative model that uses class-semantics to not only generate the features but also to discriminatively separate them. Further, using a unified model, we ensure the synthesized features have high diversity that represents the intra-class differences and variable localization precision in the detected bounding boxes. We test our approach on three object detection benchmarks, PASCAL VOC, MSCOCO, and ILSVRC detection, under both conventional and generalized settings, showing impressive gains over the state-of-the-art methods. Our codes are available at https://github.com/nasir6/zero_shot_detection.
Towards Robust and Reproducible Active Learning Using Neural Networks
Feb 21, 2020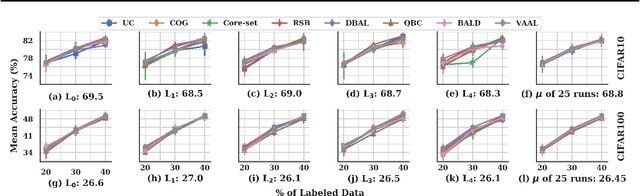
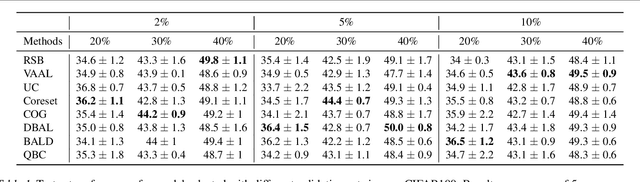
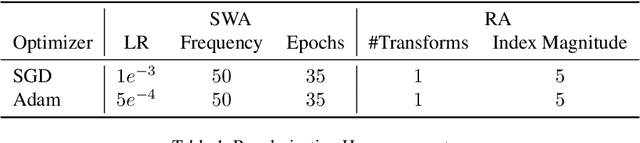
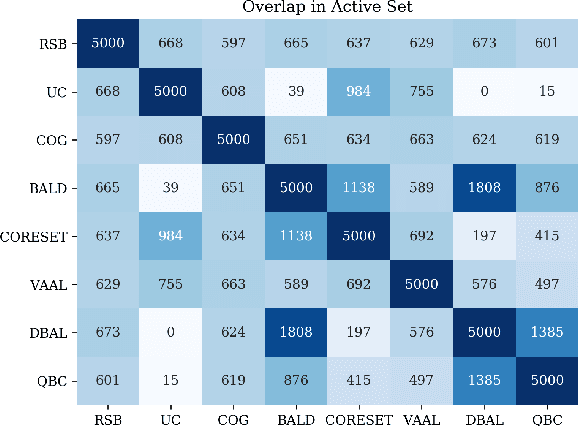
Abstract:Active learning (AL) is a promising ML paradigm that has the potential to parse through large unlabeled data and help reduce annotation cost in domains where labeling entire data can be prohibitive. Recently proposed neural network based AL methods use different heuristics to accomplish this goal. In this study, we show that recent AL methods offer a gain over random baseline under a brittle combination of experimental conditions. We demonstrate that such marginal gains vanish when experimental factors are changed, leading to reproducibility issues and suggesting that AL methods lack robustness. We also observe that with a properly tuned model, which employs recently proposed regularization techniques, the performance significantly improves for all AL methods including the random sampling baseline, and performance differences among the AL methods become negligible. Based on these observations, we suggest a set of experiments that are critical to assess the true effectiveness of an AL method. To facilitate these experiments we also present an open source toolkit. We believe our findings and recommendations will help advance reproducible research in robust AL using neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge