Ghadeer O. Ghosheh
Understanding Missingness in Time-series Electronic Health Records for Individualized Representation
Feb 24, 2024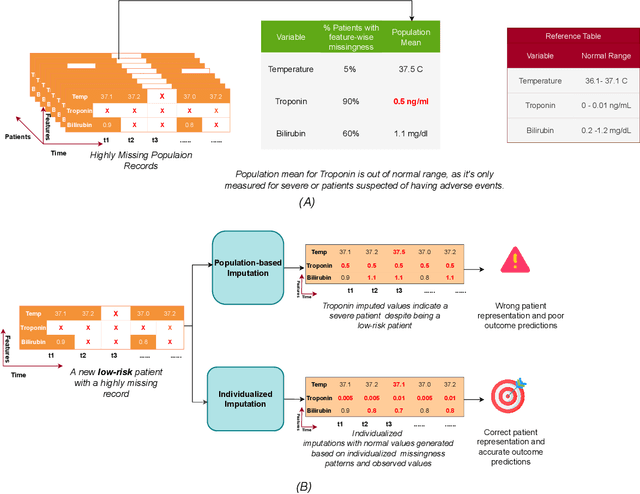
Abstract:With the widespread of machine learning models for healthcare applications, there is increased interest in building applications for personalized medicine. Despite the plethora of proposed research for personalized medicine, very few focus on representing missingness and learning from the missingness patterns in time-series Electronic Health Records (EHR) data. The lack of focus on missingness representation in an individualized way limits the full utilization of machine learning applications towards true personalization. In this brief communication, we highlight new insights into patterns of missingness with real-world examples and implications of missingness in EHRs. The insights in this work aim to bridge the gap between theoretical assumptions and practical observations in real-world EHRs. We hope this work will open new doors for exploring directions for better representation in predictive modelling for true personalization.
A Perspective on Individualized Treatment Effects Estimation from Time-series Health Data
Feb 07, 2024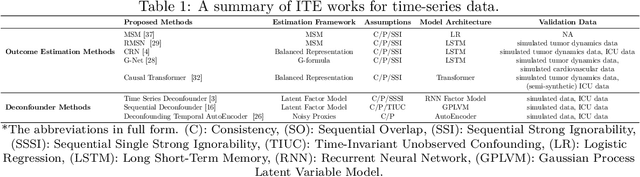
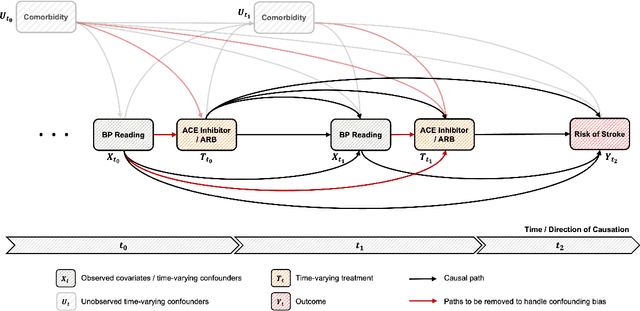
Abstract:The burden of diseases is rising worldwide, with unequal treatment efficacy for patient populations that are underrepresented in clinical trials. Healthcare, however, is driven by the average population effect of medical treatments and, therefore, operates in a "one-size-fits-all" approach, not necessarily what best fits each patient. These facts suggest a pressing need for methodologies to study individualized treatment effects (ITE) to drive personalized treatment. Despite the increased interest in machine-learning-driven ITE estimation models, the vast majority focus on tabular data with limited review and understanding of methodologies proposed for time-series electronic health records (EHRs). To this end, this work provides an overview of ITE works for time-series data and insights into future research. The work summarizes the latest work in the literature and reviews it in light of theoretical assumptions, types of treatment settings, and computational frameworks. Furthermore, this work discusses challenges and future research directions for ITEs in a time-series setting. We hope this work opens new directions and serves as a resource for understanding one of the exciting yet under-studied research areas.
IGNITE: Individualized GeNeration of Imputations in Time-series Electronic health records
Jan 09, 2024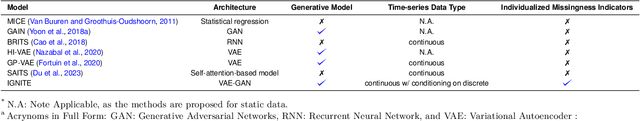
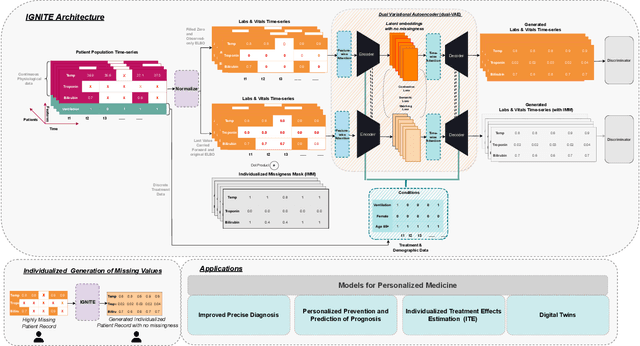
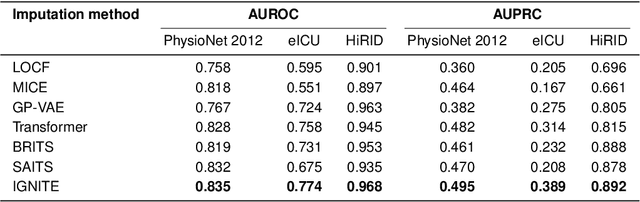
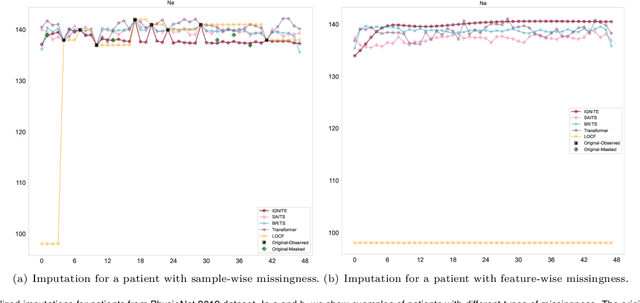
Abstract:Electronic Health Records present a valuable modality for driving personalized medicine, where treatment is tailored to fit individual-level differences. For this purpose, many data-driven machine learning and statistical models rely on the wealth of longitudinal EHRs to study patients' physiological and treatment effects. However, longitudinal EHRs tend to be sparse and highly missing, where missingness could also be informative and reflect the underlying patient's health status. Therefore, the success of data-driven models for personalized medicine highly depends on how the EHR data is represented from physiological data, treatments, and the missing values in the data. To this end, we propose a novel deep-learning model that learns the underlying patient dynamics over time across multivariate data to generate personalized realistic values conditioning on an individual's demographic characteristics and treatments. Our proposed model, IGNITE (Individualized GeNeration of Imputations in Time-series Electronic health records), utilises a conditional dual-variational autoencoder augmented with dual-stage attention to generate missing values for an individual. In IGNITE, we further propose a novel individualized missingness mask (IMM), which helps our model generate values based on the individual's observed data and missingness patterns. We further extend the use of IGNITE from imputing missingness to a personalized data synthesizer, where it generates missing EHRs that were never observed prior or even generates new patients for various applications. We validate our model on three large publicly available datasets and show that IGNITE outperforms state-of-the-art approaches in missing data reconstruction and task prediction.
Synthesizing Mixed-type Electronic Health Records using Diffusion Models
Feb 28, 2023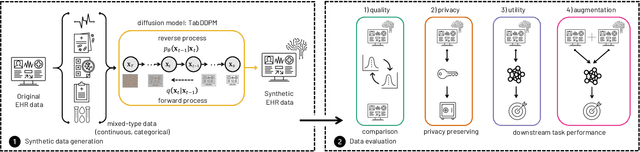
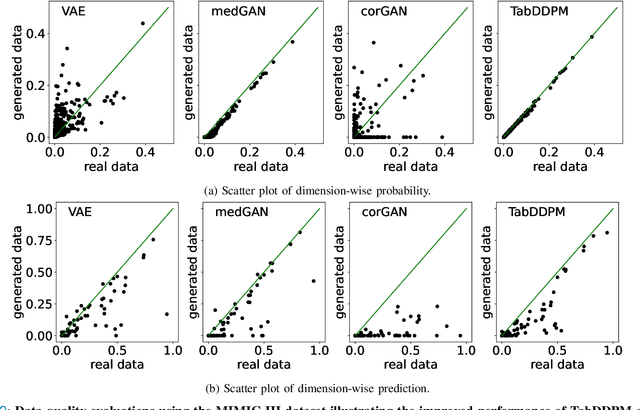
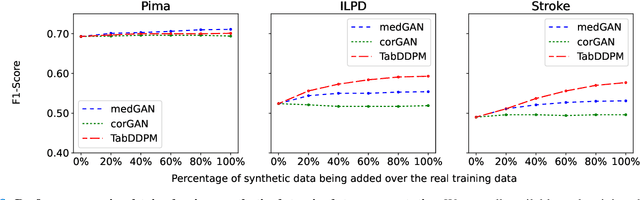
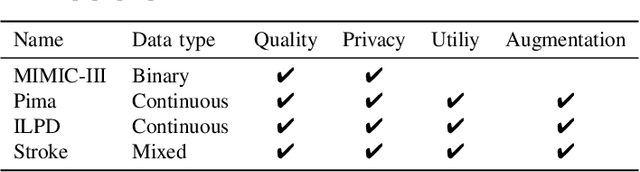
Abstract:Electronic Health Records (EHRs) contain sensitive patient information, which presents privacy concerns when sharing such data. Synthetic data generation is a promising solution to mitigate these risks, often relying on deep generative models such as Generative Adversarial Networks (GANs). However, recent studies have shown that diffusion models offer several advantages over GANs, such as generation of more realistic synthetic data and stable training in generating data modalities, including image, text, and sound. In this work, we investigate the potential of diffusion models for generating realistic mixed-type tabular EHRs, comparing TabDDPM model with existing methods on four datasets in terms of data quality, utility, privacy, and augmentation. Our experiments demonstrate that TabDDPM outperforms the state-of-the-art models across all evaluation metrics, except for privacy, which confirms the trade-off between privacy and utility.
Clinical prediction system of complications among COVID-19 patients: a development and validation retrospective multicentre study
Nov 28, 2020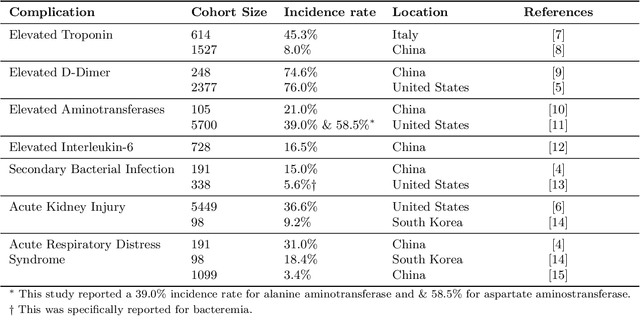


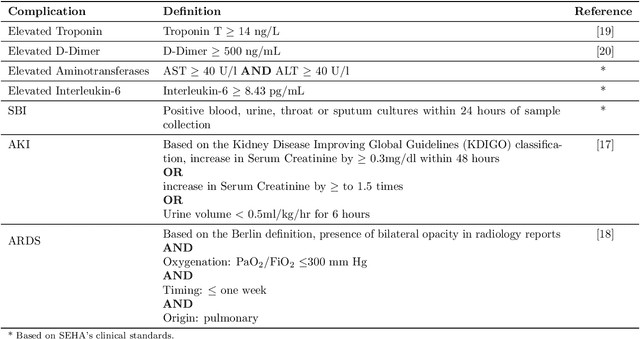
Abstract:Existing prognostic tools mainly focus on predicting the risk of mortality among patients with coronavirus disease 2019. However, clinical evidence suggests that COVID-19 can result in non-mortal complications that affect patient prognosis. To support patient risk stratification, we aimed to develop a prognostic system that predicts complications common to COVID-19. In this retrospective study, we used data collected from 3,352 COVID-19 patient encounters admitted to 18 facilities between April 1 and April 30, 2020, in Abu Dhabi (AD), UAE. The hospitals were split based on geographical proximity to assess for our proposed system's learning generalizability, AD Middle region and AD Western & Eastern regions, A and B, respectively. Using data collected during the first 24 hours of admission, the machine learning-based prognostic system predicts the risk of developing any of seven complications during the hospital stay. The complications include secondary bacterial infection, AKI, ARDS, and elevated biomarkers linked to increased patient severity, including d-dimer, interleukin-6, aminotransferases, and troponin. During training, the system applies an exclusion criteria, hyperparameter tuning, and model selection for each complication-specific model. The system achieves good accuracy across all complications and both regions. In test set A (587 patient encounters), the system achieves 0.91 AUROC for AKI and >0.80 AUROC for most of the other complications. In test set B (225 patient encounters), the respective system achieves 0.90 AUROC for AKI, elevated troponin, and elevated interleukin-6, and >0.80 AUROC for most of the other complications. The best performing models, as selected by our system, were mainly gradient boosting models and logistic regression. Our results show that a data-driven approach using machine learning can predict the risk of such complications with high accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge