David A. Clifton
Department of Engineering Science, University of Oxford, Oxford, UK, Oxford Suzhou Centre for Advanced Research, University of Oxford, Suzhou, Jiangsu, China
Semantic Self-Distillation for Language Model Uncertainty
Feb 04, 2026Abstract:Large language models present challenges for principled uncertainty quantification, in part due to their complexity and the diversity of their outputs. Semantic dispersion, or the variance in the meaning of sampled answers, has been proposed as a useful proxy for model uncertainty, but the associated computational cost prohibits its use in latency-critical applications. We show that sampled semantic distributions can be distilled into lightweight student models which estimate a prompt-conditioned uncertainty before the language model generates an answer token. The student model predicts a semantic distribution over possible answers; the entropy of this distribution provides an effective uncertainty signal for hallucination prediction, and the probability density allows candidate answers to be evaluated for reliability. On TriviaQA, our student models match or outperform finite-sample semantic dispersion for hallucination prediction and provide a strong signal for out-of-domain answer detection. We term this technique Semantic Self-Distillation (SSD), which we suggest provides a general framework for distilling predictive uncertainty in complex output spaces beyond language.
Autonomous Knowledge Graph Exploration with Adaptive Breadth-Depth Retrieval
Jan 20, 2026Abstract:Retrieving evidence for language model queries from knowledge graphs requires balancing broad search across the graph with multi-hop traversal to follow relational links. Similarity-based retrievers provide coverage but remain shallow, whereas traversal-based methods rely on selecting seed nodes to start exploration, which can fail when queries span multiple entities and relations. We introduce ARK: Adaptive Retriever of Knowledge, an agentic KG retriever that gives a language model control over this breadth-depth tradeoff using a two-operation toolset: global lexical search over node descriptors and one-hop neighborhood exploration that composes into multi-hop traversal. ARK alternates between breadth-oriented discovery and depth-oriented expansion without depending on a fragile seed selection, a pre-set hop depth, or requiring retrieval training. ARK adapts tool use to queries, using global search for language-heavy queries and neighborhood exploration for relation-heavy queries. On STaRK, ARK reaches 59.1% average Hit@1 and 67.4 average MRR, improving average Hit@1 by up to 31.4% and average MRR by up to 28.0% over retrieval-based and agentic training-free methods. Finally, we distill ARK's tool-use trajectories from a large teacher into an 8B model via label-free imitation, improving Hit@1 by +7.0, +26.6, and +13.5 absolute points over the base 8B model on AMAZON, MAG, and PRIME datasets, respectively, while retaining up to 98.5% of the teacher's Hit@1 rate.
Graph AI generates neurological hypotheses validated in molecular, organoid, and clinical systems
Dec 13, 2025Abstract:Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
MARS2 2025 Challenge on Multimodal Reasoning: Datasets, Methods, Results, Discussion, and Outlook
Sep 17, 2025
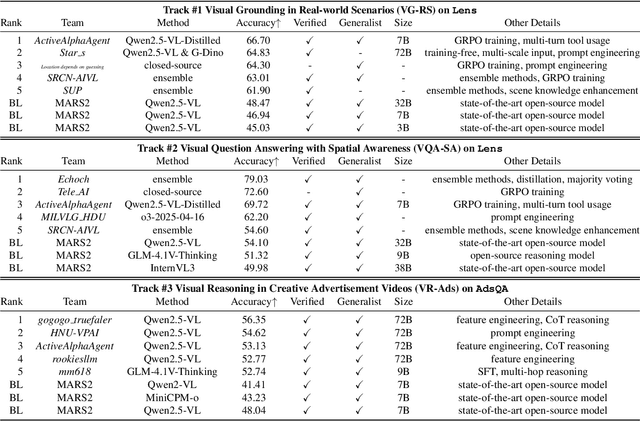

Abstract:This paper reviews the MARS2 2025 Challenge on Multimodal Reasoning. We aim to bring together different approaches in multimodal machine learning and LLMs via a large benchmark. We hope it better allows researchers to follow the state-of-the-art in this very dynamic area. Meanwhile, a growing number of testbeds have boosted the evolution of general-purpose large language models. Thus, this year's MARS2 focuses on real-world and specialized scenarios to broaden the multimodal reasoning applications of MLLMs. Our organizing team released two tailored datasets Lens and AdsQA as test sets, which support general reasoning in 12 daily scenarios and domain-specific reasoning in advertisement videos, respectively. We evaluated 40+ baselines that include both generalist MLLMs and task-specific models, and opened up three competition tracks, i.e., Visual Grounding in Real-world Scenarios (VG-RS), Visual Question Answering with Spatial Awareness (VQA-SA), and Visual Reasoning in Creative Advertisement Videos (VR-Ads). Finally, 76 teams from the renowned academic and industrial institutions have registered and 40+ valid submissions (out of 1200+) have been included in our ranking lists. Our datasets, code sets (40+ baselines and 15+ participants' methods), and rankings are publicly available on the MARS2 workshop website and our GitHub organization page https://github.com/mars2workshop/, where our updates and announcements of upcoming events will be continuously provided.
RiskAgent: Autonomous Medical AI Copilot for Generalist Risk Prediction
Mar 05, 2025Abstract:The application of Large Language Models (LLMs) to various clinical applications has attracted growing research attention. However, real-world clinical decision-making differs significantly from the standardized, exam-style scenarios commonly used in current efforts. In this paper, we present the RiskAgent system to perform a broad range of medical risk predictions, covering over 387 risk scenarios across diverse complex diseases, e.g., cardiovascular disease and cancer. RiskAgent is designed to collaborate with hundreds of clinical decision tools, i.e., risk calculators and scoring systems that are supported by evidence-based medicine. To evaluate our method, we have built the first benchmark MedRisk specialized for risk prediction, including 12,352 questions spanning 154 diseases, 86 symptoms, 50 specialties, and 24 organ systems. The results show that our RiskAgent, with 8 billion model parameters, achieves 76.33% accuracy, outperforming the most recent commercial LLMs, o1, o3-mini, and GPT-4.5, and doubling the 38.39% accuracy of GPT-4o. On rare diseases, e.g., Idiopathic Pulmonary Fibrosis (IPF), RiskAgent outperforms o1 and GPT-4.5 by 27.27% and 45.46% accuracy, respectively. Finally, we further conduct a generalization evaluation on an external evidence-based diagnosis benchmark and show that our RiskAgent achieves the best results. These encouraging results demonstrate the great potential of our solution for diverse diagnosis domains. To improve the adaptability of our model in different scenarios, we have built and open-sourced a family of models ranging from 1 billion to 70 billion parameters. Our code, data, and models are all available at https://github.com/AI-in-Health/RiskAgent.
Individualised Treatment Effects Estimation with Composite Treatments and Composite Outcomes
Feb 12, 2025



Abstract:Estimating individualised treatment effect (ITE) -- that is the causal effect of a set of variables (also called exposures, treatments, actions, policies, or interventions), referred to as \textit{composite treatments}, on a set of outcome variables of interest, referred to as \textit{composite outcomes}, for a unit from observational data -- remains a fundamental problem in causal inference with applications across disciplines, such as healthcare, economics, education, social science, marketing, and computer science. Previous work in causal machine learning for ITE estimation is limited to simple settings, like single treatments and single outcomes. This hinders their use in complex real-world scenarios; for example, consider studying the effect of different ICU interventions, such as beta-blockers and statins for a patient admitted for heart surgery, on different outcomes of interest such as atrial fibrillation and in-hospital mortality. The limited research into composite treatments and outcomes is primarily due to data scarcity for all treatments and outcomes. To address the above challenges, we propose a novel and innovative hypernetwork-based approach, called \emph{H-Learner}, to solve ITE estimation under composite treatments and composite outcomes, which tackles the data scarcity issue by dynamically sharing information across treatments and outcomes. Our empirical analysis with binary and arbitrary composite treatments and outcomes demonstrates the effectiveness of the proposed approach compared to existing methods.
Exploring the latent space of diffusion models directly through singular value decomposition
Feb 04, 2025Abstract:Despite the groundbreaking success of diffusion models in generating high-fidelity images, their latent space remains relatively under-explored, even though it holds significant promise for enabling versatile and interpretable image editing capabilities. The complicated denoising trajectory and high dimensionality of the latent space make it extremely challenging to interpret. Existing methods mainly explore the feature space of U-Net in Diffusion Models (DMs) instead of the latent space itself. In contrast, we directly investigate the latent space via Singular Value Decomposition (SVD) and discover three useful properties that can be used to control generation results without the requirements of data collection and maintain identity fidelity generated images. Based on these properties, we propose a novel image editing framework that is capable of learning arbitrary attributes from one pair of latent codes destined by text prompts in Stable Diffusion Models. To validate our approach, extensive experiments are conducted to demonstrate its effectiveness and flexibility in image editing. We will release our codes soon to foster further research and applications in this area.
Enhancing Generalization via Sharpness-Aware Trajectory Matching for Dataset Condensation
Feb 03, 2025



Abstract:Dataset condensation aims to synthesize datasets with a few representative samples that can effectively represent the original datasets. This enables efficient training and produces models with performance close to those trained on the original sets. Most existing dataset condensation methods conduct dataset learning under the bilevel (inner- and outer-loop) based optimization. However, the preceding methods perform with limited dataset generalization due to the notoriously complicated loss landscape and expensive time-space complexity of the inner-loop unrolling of bilevel optimization. These issues deteriorate when the datasets are learned via matching the trajectories of networks trained on the real and synthetic datasets with a long horizon inner-loop. To address these issues, we introduce Sharpness-Aware Trajectory Matching (SATM), which enhances the generalization capability of learned synthetic datasets by optimising the sharpness of the loss landscape and objective simultaneously. Moreover, our approach is coupled with an efficient hypergradient approximation that is mathematically well-supported and straightforward to implement along with controllable computational overhead. Empirical evaluations of SATM demonstrate its effectiveness across various applications, including in-domain benchmarks and out-of-domain settings. Moreover, its easy-to-implement properties afford flexibility, allowing it to integrate with other advanced sharpness-aware minimizers. Our code will be released.
Continuous Knowledge-Preserving Decomposition for Few-Shot Continual Learning
Jan 09, 2025



Abstract:Few-shot class-incremental learning (FSCIL) involves learning new classes from limited data while retaining prior knowledge, and often results in catastrophic forgetting. Existing methods either freeze backbone networks to preserve knowledge, which limits adaptability, or rely on additional modules or prompts, introducing inference overhead. To this end, we propose Continuous Knowledge-Preserving Decomposition for FSCIL (CKPD-FSCIL), a framework that decomposes a model's weights into two parts: one that compacts existing knowledge (knowledge-sensitive components) and another that carries redundant capacity to accommodate new abilities (redundant-capacity components). The decomposition is guided by a covariance matrix from replay samples, ensuring principal components align with classification abilities. During adaptation, we freeze the knowledge-sensitive components and only adapt the redundant-capacity components, fostering plasticity while minimizing interference without changing the architecture or increasing overhead. Additionally, CKPD introduces an adaptive layer selection strategy to identify layers with redundant capacity, dynamically allocating adapters. Experiments on multiple benchmarks show that CKPD-FSCIL outperforms state-of-the-art methods.
Enhancing Online Continual Learning with Plug-and-Play State Space Model and Class-Conditional Mixture of Discretization
Dec 24, 2024



Abstract:Online continual learning (OCL) seeks to learn new tasks from data streams that appear only once, while retaining knowledge of previously learned tasks. Most existing methods rely on replay, focusing on enhancing memory retention through regularization or distillation. However, they often overlook the adaptability of the model, limiting the ability to learn generalizable and discriminative features incrementally from online training data. To address this, we introduce a plug-and-play module, S6MOD, which can be integrated into most existing methods and directly improve adaptability. Specifically, S6MOD introduces an extra branch after the backbone, where a mixture of discretization selectively adjusts parameters in a selective state space model, enriching selective scan patterns such that the model can adaptively select the most sensitive discretization method for current dynamics. We further design a class-conditional routing algorithm for dynamic, uncertainty-based adjustment and implement a contrastive discretization loss to optimize it. Extensive experiments combining our module with various models demonstrate that S6MOD significantly enhances model adaptability, leading to substantial performance gains and achieving the state-of-the-art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge