Sihao Liu
Daniel
KGCE: Knowledge-Augmented Dual-Graph Evaluator for Cross-Platform Educational Agent Benchmarking with Multimodal Language Models
Jan 04, 2026Abstract:With the rapid adoption of multimodal large language models (MLMs) in autonomous agents, cross-platform task execution capabilities in educational settings have garnered significant attention. However, existing benchmark frameworks still exhibit notable deficiencies in supporting cross-platform tasks in educational contexts, especially when dealing with school-specific software (such as XiaoYa Intelligent Assistant, HuaShi XiaZi, etc.), where the efficiency of agents often significantly decreases due to a lack of understanding of the structural specifics of these private-domain software. Additionally, current evaluation methods heavily rely on coarse-grained metrics like goal orientation or trajectory matching, making it challenging to capture the detailed execution and efficiency of agents in complex tasks. To address these issues, we propose KGCE (Knowledge-Augmented Dual-Graph Evaluator for Cross-Platform Educational Agent Benchmarking with Multimodal Language Models), a novel benchmarking platform that integrates knowledge base enhancement and a dual-graph evaluation framework. We first constructed a dataset comprising 104 education-related tasks, covering Windows, Android, and cross-platform collaborative tasks. KGCE introduces a dual-graph evaluation framework that decomposes tasks into multiple sub-goals and verifies their completion status, providing fine-grained evaluation metrics. To overcome the execution bottlenecks of existing agents in private-domain tasks, we developed an enhanced agent system incorporating a knowledge base specific to school-specific software. The code can be found at https://github.com/Kinginlife/KGCE.
Dynamic Context-oriented Decomposition for Task-aware Low-rank Adaptation with Less Forgetting and Faster Convergence
Jun 16, 2025



Abstract:Conventional low-rank adaptation methods build adapters without considering data context, leading to sub-optimal fine-tuning performance and severe forgetting of inherent world knowledge. In this paper, we propose context-oriented decomposition adaptation (CorDA), a novel method that initializes adapters in a task-aware manner. Concretely, we develop context-oriented singular value decomposition, where we collect covariance matrices of input activations for each linear layer using sampled data from the target task, and apply SVD to the product of weight matrix and its corresponding covariance matrix. By doing so, the task-specific capability is compacted into the principal components. Thanks to the task awareness, our method enables two optional adaptation modes, knowledge-preserved mode (KPM) and instruction-previewed mode (IPM), providing flexibility to choose between freezing the principal components to preserve their associated knowledge or adapting them to better learn a new task. We further develop CorDA++ by deriving a metric that reflects the compactness of task-specific principal components, and then introducing dynamic covariance selection and dynamic rank allocation strategies based on the same metric. The two strategies provide each layer with the most representative covariance matrix and a proper rank allocation. Experimental results show that CorDA++ outperforms CorDA by a significant margin. CorDA++ in KPM not only achieves better fine-tuning performance than LoRA, but also mitigates the forgetting of pre-trained knowledge in both large language models and vision language models. For IPM, our method exhibits faster convergence, \emph{e.g.,} 4.5x speedup over QLoRA, and improves adaptation performance in various scenarios, outperforming strong baseline methods. Our method has been integrated into the PEFT library developed by Hugging Face.
Enhancing Online Continual Learning with Plug-and-Play State Space Model and Class-Conditional Mixture of Discretization
Dec 24, 2024



Abstract:Online continual learning (OCL) seeks to learn new tasks from data streams that appear only once, while retaining knowledge of previously learned tasks. Most existing methods rely on replay, focusing on enhancing memory retention through regularization or distillation. However, they often overlook the adaptability of the model, limiting the ability to learn generalizable and discriminative features incrementally from online training data. To address this, we introduce a plug-and-play module, S6MOD, which can be integrated into most existing methods and directly improve adaptability. Specifically, S6MOD introduces an extra branch after the backbone, where a mixture of discretization selectively adjusts parameters in a selective state space model, enriching selective scan patterns such that the model can adaptively select the most sensitive discretization method for current dynamics. We further design a class-conditional routing algorithm for dynamic, uncertainty-based adjustment and implement a contrastive discretization loss to optimize it. Extensive experiments combining our module with various models demonstrate that S6MOD significantly enhances model adaptability, leading to substantial performance gains and achieving the state-of-the-art results.
FlexiDrop: Theoretical Insights and Practical Advances in Random Dropout Method on GNNs
May 30, 2024



Abstract:Graph Neural Networks (GNNs) are powerful tools for handling graph-type data. Recently, GNNs have been widely applied in various domains, but they also face some issues, such as overfitting, over-smoothing and non-robustness. The existing research indicates that random dropout methods are an effective way to address these issues. However, random dropout methods in GNNs still face unresolved problems. Currently, the choice of dropout rate, often determined by heuristic or grid search methods, can increase the generalization error, contradicting the principal aims of dropout. In this paper, we propose a novel random dropout method for GNNs called FlexiDrop. First, we conduct a theoretical analysis of dropout in GNNs using rademacher complexity and demonstrate that the generalization error of traditional random dropout methods is constrained by a function related to the dropout rate. Subsequently, we use this function as a regularizer to unify the dropout rate and empirical loss within a single loss function, optimizing them simultaneously. Therefore, our method enables adaptive adjustment of the dropout rate and theoretically balances the trade-off between model complexity and generalization ability. Furthermore, extensive experimental results on benchmark datasets show that FlexiDrop outperforms traditional random dropout methods in GNNs.
Gaussian Control with Hierarchical Semantic Graphs in 3D Human Recovery
May 21, 2024



Abstract:Although 3D Gaussian Splatting (3DGS) has recently made progress in 3D human reconstruction, it primarily relies on 2D pixel-level supervision, overlooking the geometric complexity and topological relationships of different body parts. To address this gap, we introduce the Hierarchical Graph Human Gaussian Control (HUGS) framework for achieving high-fidelity 3D human reconstruction. Our approach involves leveraging explicitly semantic priors of body parts to ensure the consistency of geometric topology, thereby enabling the capture of the complex geometrical and topological associations among body parts. Additionally, we disentangle high-frequency features from global human features to refine surface details in body parts. Extensive experiments demonstrate that our method exhibits superior performance in human body reconstruction, particularly in enhancing surface details and accurately reconstructing body part junctions. Codes are available at https://wanghongsheng01.github.io/HUGS/.
Functional Connectome: Approximating Brain Networks with Artificial Neural Networks
Nov 23, 2022
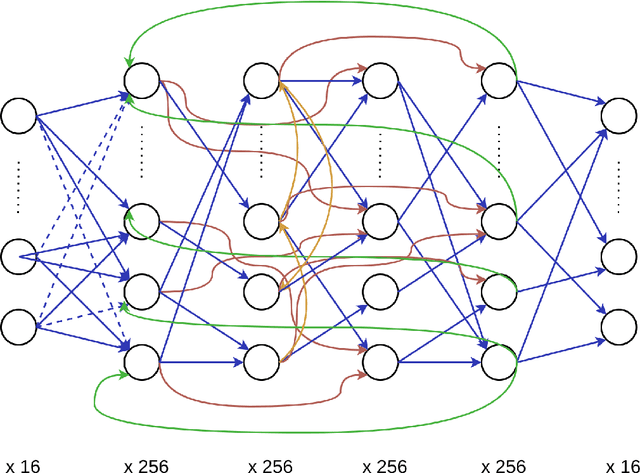


Abstract:We aimed to explore the capability of deep learning to approximate the function instantiated by biological neural circuits-the functional connectome. Using deep neural networks, we performed supervised learning with firing rate observations drawn from synthetically constructed neural circuits, as well as from an empirically supported Boundary Vector Cell-Place Cell network. The performance of trained networks was quantified using a range of criteria and tasks. Our results show that deep neural networks were able to capture the computations performed by synthetic biological networks with high accuracy, and were highly data efficient and robust to biological plasticity. We show that trained deep neural networks are able to perform zero-shot generalisation in novel environments, and allows for a wealth of tasks such as decoding the animal's location in space with high accuracy. Our study reveals a novel and promising direction in systems neuroscience, and can be expanded upon with a multitude of downstream applications, for example, goal-directed reinforcement learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge