Krzysztof J. Geras
A Multi-Modal AI System for Screening Mammography: Integrating 2D and 3D Imaging to Improve Breast Cancer Detection in a Prospective Clinical Study
Apr 08, 2025
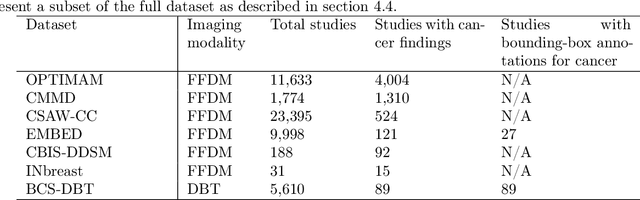
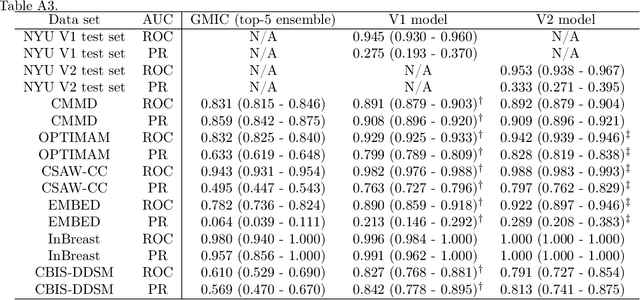
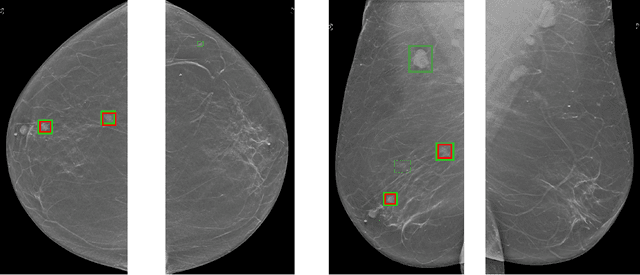
Abstract:Although digital breast tomosynthesis (DBT) improves diagnostic performance over full-field digital mammography (FFDM), false-positive recalls remain a concern in breast cancer screening. We developed a multi-modal artificial intelligence system integrating FFDM, synthetic mammography, and DBT to provide breast-level predictions and bounding-box localizations of suspicious findings. Our AI system, trained on approximately 500,000 mammography exams, achieved 0.945 AUROC on an internal test set. It demonstrated capacity to reduce recalls by 31.7% and radiologist workload by 43.8% while maintaining 100% sensitivity, underscoring its potential to improve clinical workflows. External validation confirmed strong generalizability, reducing the gap to a perfect AUROC by 35.31%-69.14% relative to strong baselines. In prospective deployment across 18 sites, the system reduced recall rates for low-risk cases. An improved version, trained on over 750,000 exams with additional labels, further reduced the gap by 18.86%-56.62% across large external datasets. Overall, these results underscore the importance of utilizing all available imaging modalities, demonstrate the potential for clinical impact, and indicate feasibility of further reduction of the test error with increased training set when using large-capacity neural networks.
Multi-modal AI for comprehensive breast cancer prognostication
Oct 28, 2024Abstract:Treatment selection in breast cancer is guided by molecular subtypes and clinical characteristics. Recurrence risk assessment plays a crucial role in personalizing treatment. Current methods, including genomic assays, have limited accuracy and clinical utility, leading to suboptimal decisions for many patients. We developed a test for breast cancer patient stratification based on digital pathology and clinical characteristics using novel AI methods. Specifically, we utilized a vision transformer-based pan-cancer foundation model trained with self-supervised learning to extract features from digitized H&E-stained slides. These features were integrated with clinical data to form a multi-modal AI test predicting cancer recurrence and death. The test was developed and evaluated using data from a total of 8,161 breast cancer patients across 15 cohorts originating from seven countries. Of these, 3,502 patients from five cohorts were used exclusively for evaluation, while the remaining patients were used for training. Our test accurately predicted our primary endpoint, disease-free interval, in the five external cohorts (C-index: 0.71 [0.68-0.75], HR: 3.63 [3.02-4.37, p<0.01]). In a direct comparison (N=858), the AI test was more accurate than Oncotype DX, the standard-of-care 21-gene assay, with a C-index of 0.67 [0.61-0.74] versus 0.61 [0.49-0.73], respectively. Additionally, the AI test added independent information to Oncotype DX in a multivariate analysis (HR: 3.11 [1.91-5.09, p<0.01)]). The test demonstrated robust accuracy across all major breast cancer subtypes, including TNBC (C-index: 0.71 [0.62-0.81], HR: 3.81 [2.35-6.17, p=0.02]), where no diagnostic tools are currently recommended by clinical guidelines. These results suggest that our AI test can improve accuracy, extend applicability to a wider range of patients, and enhance access to treatment selection tools.
Understanding differences in applying DETR to natural and medical images
May 27, 2024Abstract:Transformer-based detectors have shown success in computer vision tasks with natural images. These models, exemplified by the Deformable DETR, are optimized through complex engineering strategies tailored to the typical characteristics of natural scenes. However, medical imaging data presents unique challenges such as extremely large image sizes, fewer and smaller regions of interest, and object classes which can be differentiated only through subtle differences. This study evaluates the applicability of these transformer-based design choices when applied to a screening mammography dataset that represents these distinct medical imaging data characteristics. Our analysis reveals that common design choices from the natural image domain, such as complex encoder architectures, multi-scale feature fusion, query initialization, and iterative bounding box refinement, do not improve and sometimes even impair object detection performance in medical imaging. In contrast, simpler and shallower architectures often achieve equal or superior results. This finding suggests that the adaptation of transformer models for medical imaging data requires a reevaluation of standard practices, potentially leading to more efficient and specialized frameworks for medical diagnosis.
Leveraging Transformers to Improve Breast Cancer Classification and Risk Assessment with Multi-modal and Longitudinal Data
Nov 15, 2023
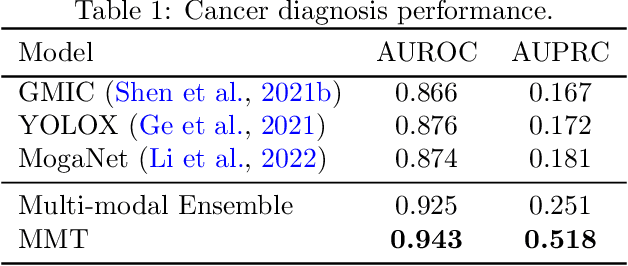
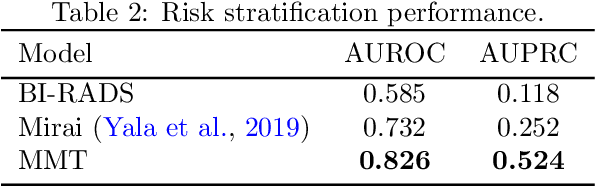
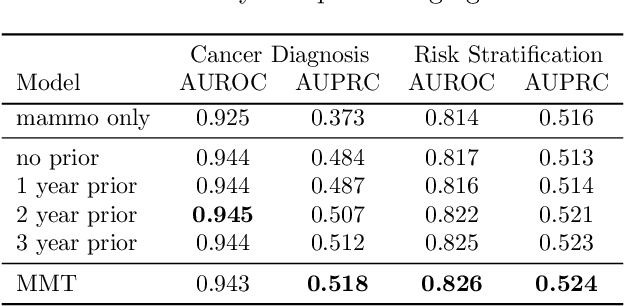
Abstract:Breast cancer screening, primarily conducted through mammography, is often supplemented with ultrasound for women with dense breast tissue. However, existing deep learning models analyze each modality independently, missing opportunities to integrate information across imaging modalities and time. In this study, we present Multi-modal Transformer (MMT), a neural network that utilizes mammography and ultrasound synergistically, to identify patients who currently have cancer and estimate the risk of future cancer for patients who are currently cancer-free. MMT aggregates multi-modal data through self-attention and tracks temporal tissue changes by comparing current exams to prior imaging. Trained on 1.3 million exams, MMT achieves an AUROC of 0.943 in detecting existing cancers, surpassing strong uni-modal baselines. For 5-year risk prediction, MMT attains an AUROC of 0.826, outperforming prior mammography-based risk models. Our research highlights the value of multi-modal and longitudinal imaging in cancer diagnosis and risk stratification.
Multiple Instance Learning via Iterative Self-Paced Supervised Contrastive Learning
Oct 17, 2022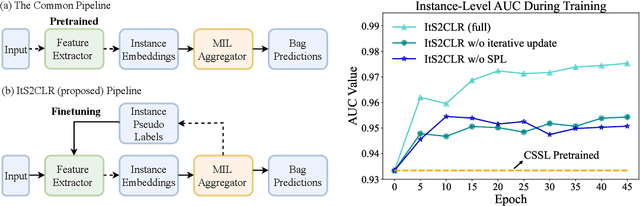
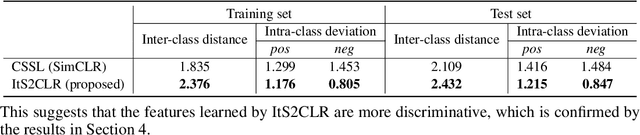
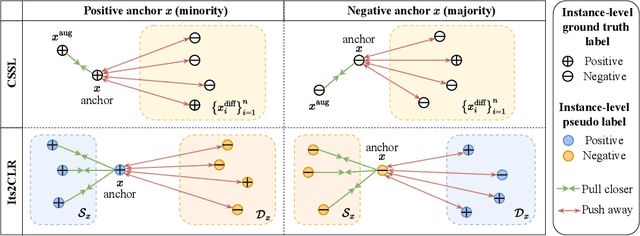

Abstract:Learning representations for individual instances when only bag-level labels are available is a fundamental challenge in multiple instance learning (MIL). Recent works have shown promising results using contrastive self-supervised learning (CSSL), which learns to push apart representations corresponding to two different randomly-selected instances. Unfortunately, in real-world applications such as medical image classification, there is often class imbalance, so randomly-selected instances mostly belong to the same majority class, which precludes CSSL from learning inter-class differences. To address this issue, we propose a novel framework, Iterative Self-paced Supervised Contrastive Learning for MIL Representations (ItS2CLR), which improves the learned representation by exploiting instance-level pseudo labels derived from the bag-level labels. The framework employs a novel self-paced sampling strategy to ensure the accuracy of pseudo labels. We evaluate ItS2CLR on three medical datasets, showing that it improves the quality of instance-level pseudo labels and representations, and outperforms existing MIL methods in terms of both bag and instance level accuracy.
3D-GMIC: an efficient deep neural network to find small objects in large 3D images
Oct 16, 2022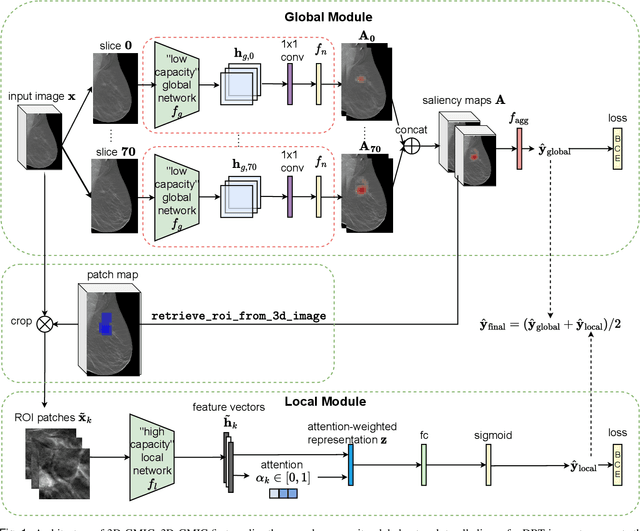
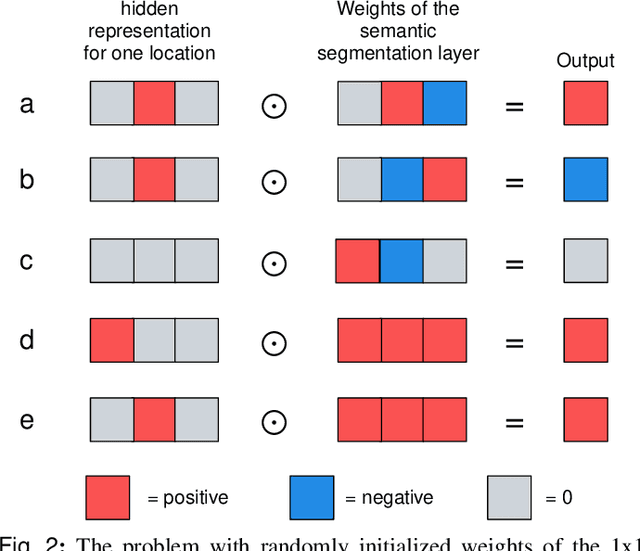
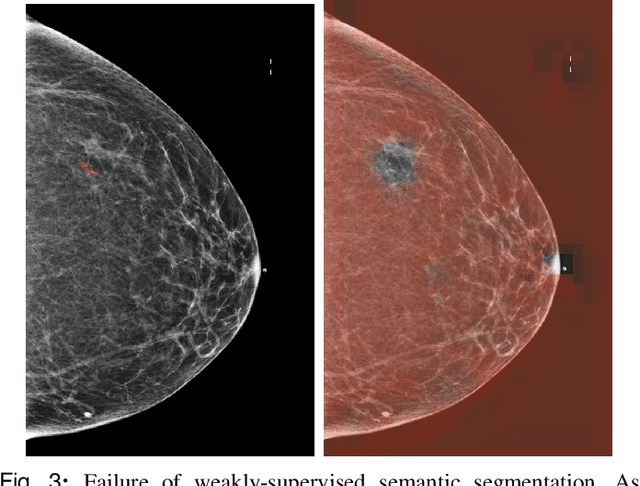
Abstract:3D imaging enables a more accurate diagnosis by providing spatial information about organ anatomy. However, using 3D images to train AI models is computationally challenging because they consist of tens or hundreds of times more pixels than their 2D counterparts. To train with high-resolution 3D images, convolutional neural networks typically resort to downsampling them or projecting them to two dimensions. In this work, we propose an effective alternative, a novel neural network architecture that enables computationally efficient classification of 3D medical images in their full resolution. Compared to off-the-shelf convolutional neural networks, 3D-GMIC uses 77.98%-90.05% less GPU memory and 91.23%-96.02% less computation. While our network is trained only with image-level labels, without segmentation labels, it explains its classification predictions by providing pixel-level saliency maps. On a dataset collected at NYU Langone Health, including 85,526 patients with full-field 2D mammography (FFDM), synthetic 2D mammography, and 3D mammography (DBT), our model, the 3D Globally-Aware Multiple Instance Classifier (3D-GMIC), achieves a breast-wise AUC of 0.831 (95% CI: 0.769-0.887) in classifying breasts with malignant findings using DBT images. As DBT and 2D mammography capture different information, averaging predictions on 2D and 3D mammography together leads to a diverse ensemble with an improved breast-wise AUC of 0.841 (95% CI: 0.768-0.895). Our model generalizes well to an external dataset from Duke University Hospital, achieving an image-wise AUC of 0.848 (95% CI: 0.798-0.896) in classifying DBT images with malignant findings.
MedFuse: Multi-modal fusion with clinical time-series data and chest X-ray images
Jul 14, 2022
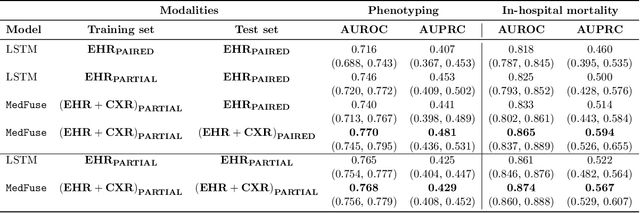

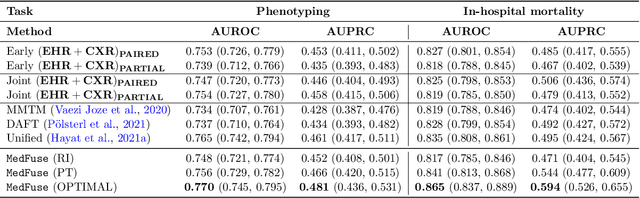
Abstract:Multi-modal fusion approaches aim to integrate information from different data sources. Unlike natural datasets, such as in audio-visual applications, where samples consist of "paired" modalities, data in healthcare is often collected asynchronously. Hence, requiring the presence of all modalities for a given sample is not realistic for clinical tasks and significantly limits the size of the dataset during training. In this paper, we propose MedFuse, a conceptually simple yet promising LSTM-based fusion module that can accommodate uni-modal as well as multi-modal input. We evaluate the fusion method and introduce new benchmark results for in-hospital mortality prediction and phenotype classification, using clinical time-series data in the MIMIC-IV dataset and corresponding chest X-ray images in MIMIC-CXR. Compared to more complex multi-modal fusion strategies, MedFuse provides a performance improvement by a large margin on the fully paired test set. It also remains robust across the partially paired test set containing samples with missing chest X-ray images. We release our code for reproducibility and to enable the evaluation of competing models in the future.
Characterizing and overcoming the greedy nature of learning in multi-modal deep neural networks
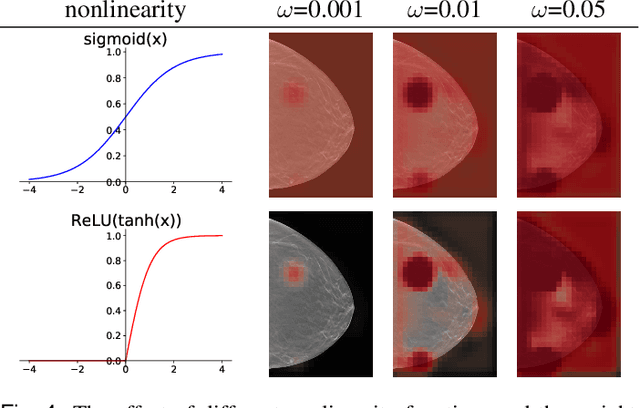
Feb 10, 2022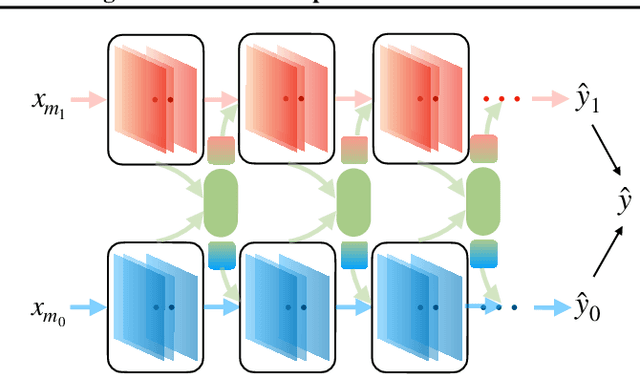
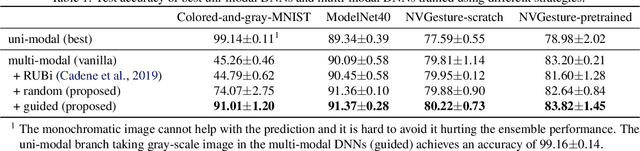
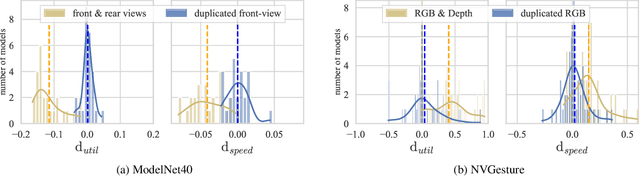
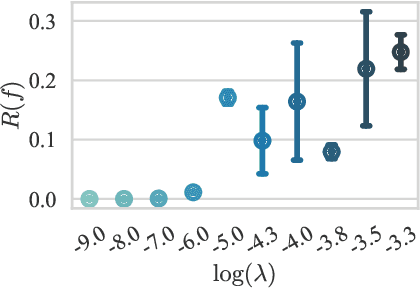
Abstract:We hypothesize that due to the greedy nature of learning in multi-modal deep neural networks, these models tend to rely on just one modality while under-fitting the other modalities. Such behavior is counter-intuitive and hurts the models' generalization, as we observe empirically. To estimate the model's dependence on each modality, we compute the gain on the accuracy when the model has access to it in addition to another modality. We refer to this gain as the conditional utilization rate. In the experiments, we consistently observe an imbalance in conditional utilization rates between modalities, across multiple tasks and architectures. Since conditional utilization rate cannot be computed efficiently during training, we introduce a proxy for it based on the pace at which the model learns from each modality, which we refer to as the conditional learning speed. We propose an algorithm to balance the conditional learning speeds between modalities during training and demonstrate that it indeed addresses the issue of greedy learning. The proposed algorithm improves the model's generalization on three datasets: Colored MNIST, Princeton ModelNet40, and NVIDIA Dynamic Hand Gesture.
Towards dynamic multi-modal phenotyping using chest radiographs and physiological data
Nov 04, 2021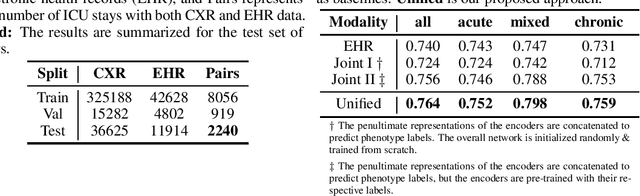
Abstract:The healthcare domain is characterized by heterogeneous data modalities, such as imaging and physiological data. In practice, the variety of medical data assists clinicians in decision-making. However, most of the current state-of-the-art deep learning models solely rely upon carefully curated data of a single modality. In this paper, we propose a dynamic training approach to learn modality-specific data representations and to integrate auxiliary features, instead of solely relying on a single modality. Our preliminary experiments results for a patient phenotyping task using physiological data in MIMIC-IV & chest radiographs in the MIMIC- CXR dataset show that our proposed approach achieves the highest area under the receiver operating characteristic curve (AUROC) (0.764 AUROC) compared to the performance of the benchmark method in previous work, which only used physiological data (0.740 AUROC). For a set of five recurring or chronic diseases with periodic acute episodes, including cardiac dysrhythmia, conduction disorders, and congestive heart failure, the AUROC improves from 0.747 to 0.798. This illustrates the benefit of leveraging the chest imaging modality in the phenotyping task and highlights the potential of multi-modal learning in medical applications.
Meta-repository of screening mammography classifiers
Aug 10, 2021
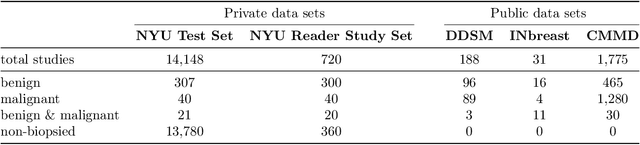
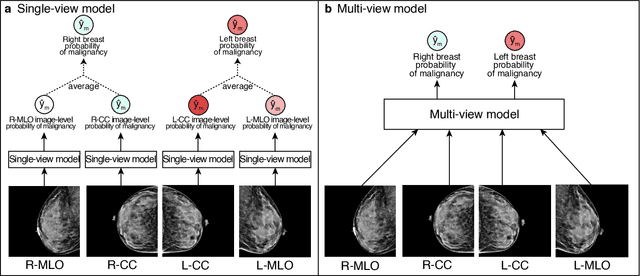
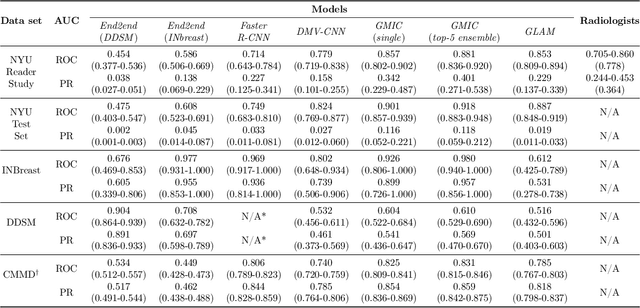
Abstract:Artificial intelligence (AI) is transforming medicine and showing promise in improving clinical diagnosis. In breast cancer screening, several recent studies show that AI has the potential to improve radiologists' accuracy, subsequently helping in early cancer diagnosis and reducing unnecessary workup. As the number of proposed models and their complexity grows, it is becoming increasingly difficult to re-implement them in order to reproduce the results and to compare different approaches. To enable reproducibility of research in this application area and to enable comparison between different methods, we release a meta-repository containing deep learning models for classification of screening mammograms. This meta-repository creates a framework that enables the evaluation of machine learning models on any private or public screening mammography data set. At its inception, our meta-repository contains five state-of-the-art models with open-source implementations and cross-platform compatibility. We compare their performance on five international data sets: two private New York University breast cancer screening data sets as well as three public (DDSM, INbreast and Chinese Mammography Database) data sets. Our framework has a flexible design that can be generalized to other medical image analysis tasks. The meta-repository is available at https://www.github.com/nyukat/mammography_metarepository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge