Jungkyu Park
A Multi-Modal AI System for Screening Mammography: Integrating 2D and 3D Imaging to Improve Breast Cancer Detection in a Prospective Clinical Study
Apr 08, 2025
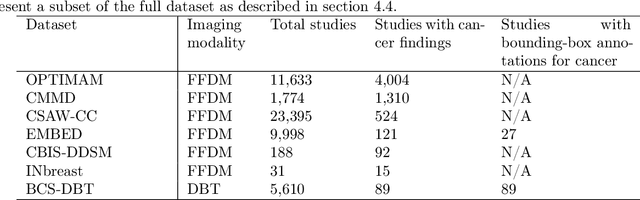
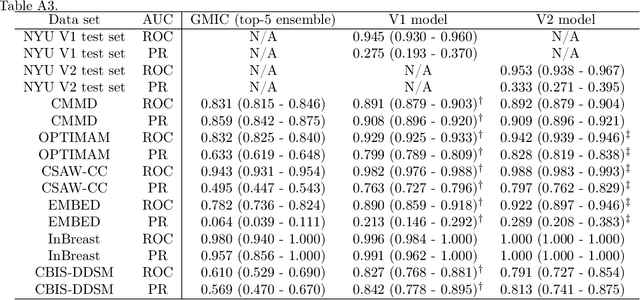
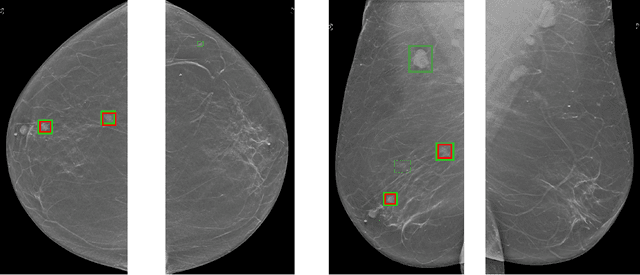
Abstract:Although digital breast tomosynthesis (DBT) improves diagnostic performance over full-field digital mammography (FFDM), false-positive recalls remain a concern in breast cancer screening. We developed a multi-modal artificial intelligence system integrating FFDM, synthetic mammography, and DBT to provide breast-level predictions and bounding-box localizations of suspicious findings. Our AI system, trained on approximately 500,000 mammography exams, achieved 0.945 AUROC on an internal test set. It demonstrated capacity to reduce recalls by 31.7% and radiologist workload by 43.8% while maintaining 100% sensitivity, underscoring its potential to improve clinical workflows. External validation confirmed strong generalizability, reducing the gap to a perfect AUROC by 35.31%-69.14% relative to strong baselines. In prospective deployment across 18 sites, the system reduced recall rates for low-risk cases. An improved version, trained on over 750,000 exams with additional labels, further reduced the gap by 18.86%-56.62% across large external datasets. Overall, these results underscore the importance of utilizing all available imaging modalities, demonstrate the potential for clinical impact, and indicate feasibility of further reduction of the test error with increased training set when using large-capacity neural networks.
A training regime to learn unified representations from complementary breast imaging modalities
Aug 16, 2024



Abstract:Full Field Digital Mammograms (FFDMs) and Digital Breast Tomosynthesis (DBT) are the two most widely used imaging modalities for breast cancer screening. Although DBT has increased cancer detection compared to FFDM, its widespread adoption in clinical practice has been slowed by increased interpretation times and a perceived decrease in the conspicuity of specific lesion types. Specifically, the non-inferiority of DBT for microcalcifications remains under debate. Due to concerns about the decrease in visual acuity, combined DBT-FFDM acquisitions remain popular, leading to overall increased exam times and radiation dosage. Enabling DBT to provide diagnostic information present in both FFDM and DBT would reduce reliance on FFDM, resulting in a reduction in both quantities. We propose a machine learning methodology that learns high-level representations leveraging the complementary diagnostic signal from both DBT and FFDM. Experiments on a large-scale data set validate our claims and show that our representations enable more accurate breast lesion detection than any DBT- or FFDM-based model.
Leveraging Transformers to Improve Breast Cancer Classification and Risk Assessment with Multi-modal and Longitudinal Data
Nov 15, 2023
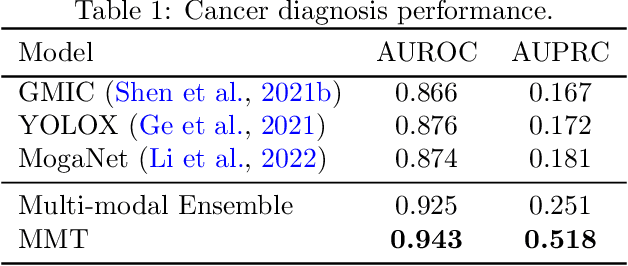
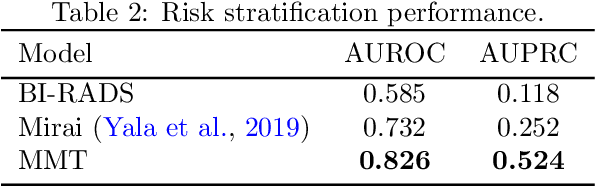
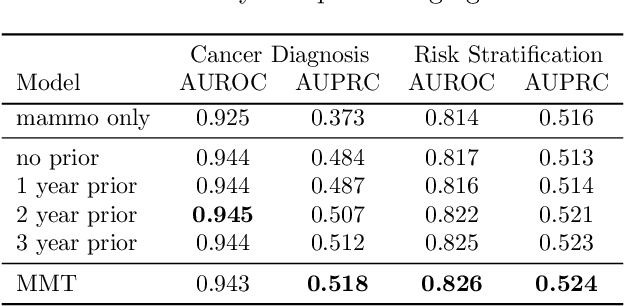
Abstract:Breast cancer screening, primarily conducted through mammography, is often supplemented with ultrasound for women with dense breast tissue. However, existing deep learning models analyze each modality independently, missing opportunities to integrate information across imaging modalities and time. In this study, we present Multi-modal Transformer (MMT), a neural network that utilizes mammography and ultrasound synergistically, to identify patients who currently have cancer and estimate the risk of future cancer for patients who are currently cancer-free. MMT aggregates multi-modal data through self-attention and tracks temporal tissue changes by comparing current exams to prior imaging. Trained on 1.3 million exams, MMT achieves an AUROC of 0.943 in detecting existing cancers, surpassing strong uni-modal baselines. For 5-year risk prediction, MMT attains an AUROC of 0.826, outperforming prior mammography-based risk models. Our research highlights the value of multi-modal and longitudinal imaging in cancer diagnosis and risk stratification.
3D-GMIC: an efficient deep neural network to find small objects in large 3D images
Oct 16, 2022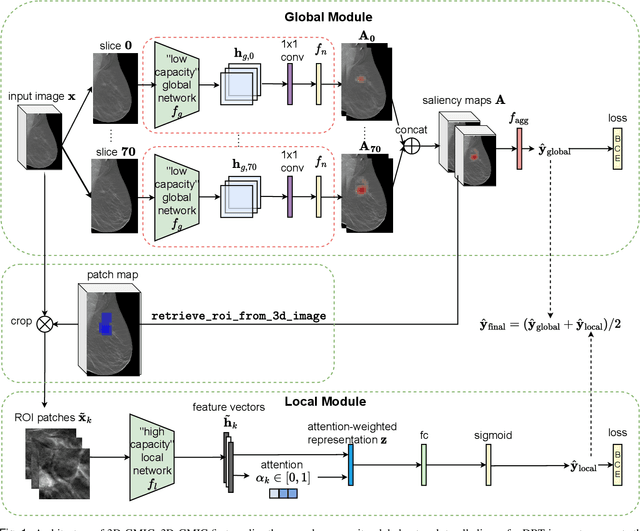
Abstract:3D imaging enables a more accurate diagnosis by providing spatial information about organ anatomy. However, using 3D images to train AI models is computationally challenging because they consist of tens or hundreds of times more pixels than their 2D counterparts. To train with high-resolution 3D images, convolutional neural networks typically resort to downsampling them or projecting them to two dimensions. In this work, we propose an effective alternative, a novel neural network architecture that enables computationally efficient classification of 3D medical images in their full resolution. Compared to off-the-shelf convolutional neural networks, 3D-GMIC uses 77.98%-90.05% less GPU memory and 91.23%-96.02% less computation. While our network is trained only with image-level labels, without segmentation labels, it explains its classification predictions by providing pixel-level saliency maps. On a dataset collected at NYU Langone Health, including 85,526 patients with full-field 2D mammography (FFDM), synthetic 2D mammography, and 3D mammography (DBT), our model, the 3D Globally-Aware Multiple Instance Classifier (3D-GMIC), achieves a breast-wise AUC of 0.831 (95% CI: 0.769-0.887) in classifying breasts with malignant findings using DBT images. As DBT and 2D mammography capture different information, averaging predictions on 2D and 3D mammography together leads to a diverse ensemble with an improved breast-wise AUC of 0.841 (95% CI: 0.768-0.895). Our model generalizes well to an external dataset from Duke University Hospital, achieving an image-wise AUC of 0.848 (95% CI: 0.798-0.896) in classifying DBT images with malignant findings.
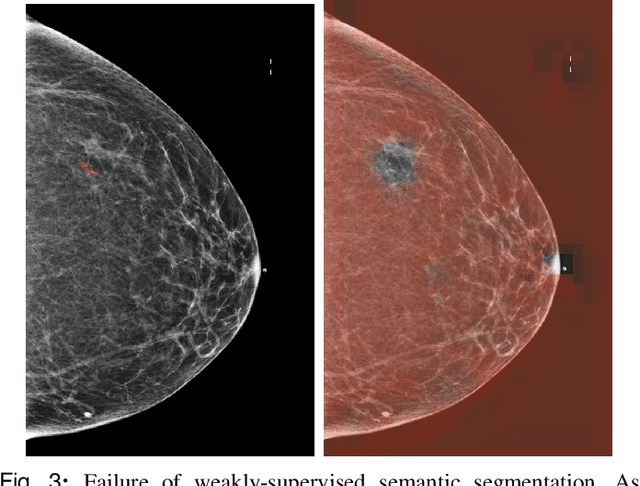
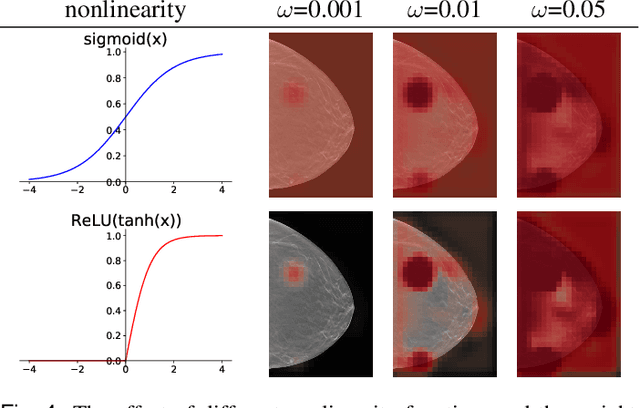
Investigating and Simplifying Masking-based Saliency Methods for Model Interpretability
Oct 19, 2020



Abstract:Saliency maps that identify the most informative regions of an image for a classifier are valuable for model interpretability. A common approach to creating saliency maps involves generating input masks that mask out portions of an image to maximally deteriorate classification performance, or mask in an image to preserve classification performance. Many variants of this approach have been proposed in the literature, such as counterfactual generation and optimizing over a Gumbel-Softmax distribution. Using a general formulation of masking-based saliency methods, we conduct an extensive evaluation study of a number of recently proposed variants to understand which elements of these methods meaningfully improve performance. Surprisingly, we find that a well-tuned, relatively simple formulation of a masking-based saliency model outperforms many more complex approaches. We find that the most important ingredients for high quality saliency map generation are (1) using both masked-in and masked-out objectives and (2) training the classifier alongside the masking model. Strikingly, we show that a masking model can be trained with as few as 10 examples per class and still generate saliency maps with only a 0.7-point increase in localization error.
Reducing false-positive biopsies with deep neural networks that utilize local and global information in screening mammograms
Sep 19, 2020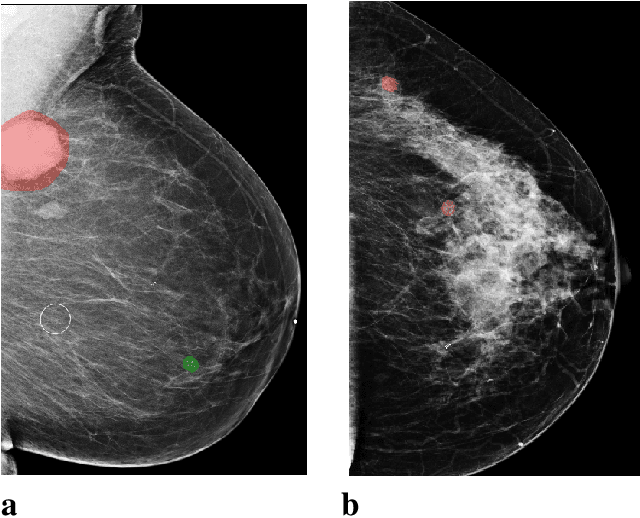
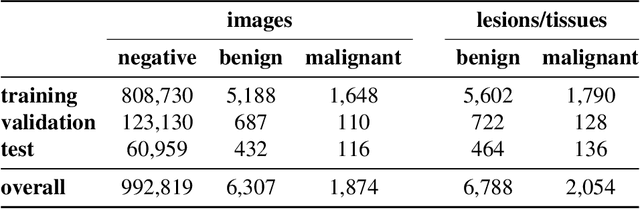
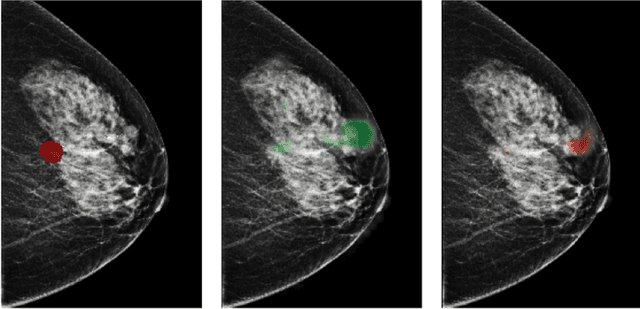
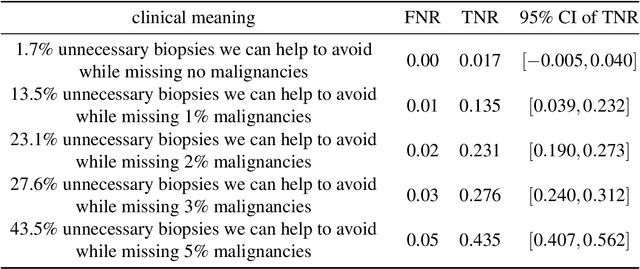
Abstract:Breast cancer is the most common cancer in women, and hundreds of thousands of unnecessary biopsies are done around the world at a tremendous cost. It is crucial to reduce the rate of biopsies that turn out to be benign tissue. In this study, we build deep neural networks (DNNs) to classify biopsied lesions as being either malignant or benign, with the goal of using these networks as second readers serving radiologists to further reduce the number of false positive findings. We enhance the performance of DNNs that are trained to learn from small image patches by integrating global context provided in the form of saliency maps learned from the entire image into their reasoning, similar to how radiologists consider global context when evaluating areas of interest. Our experiments are conducted on a dataset of 229,426 screening mammography exams from 141,473 patients. We achieve an AUC of 0.8 on a test set consisting of 464 benign and 136 malignant lesions.
An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department
Aug 04, 2020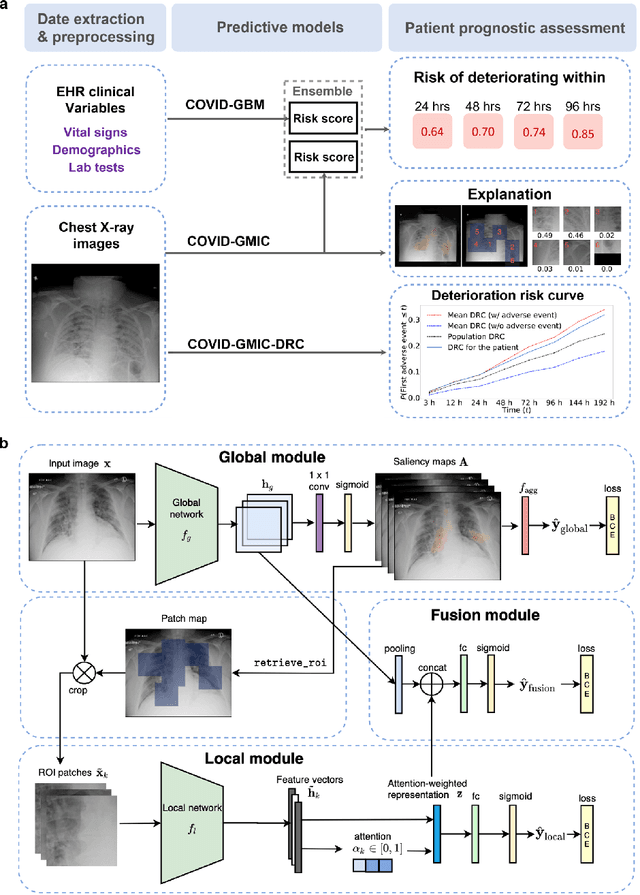
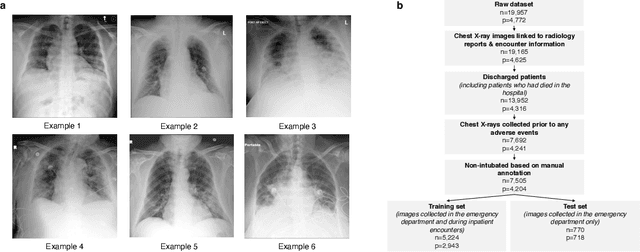
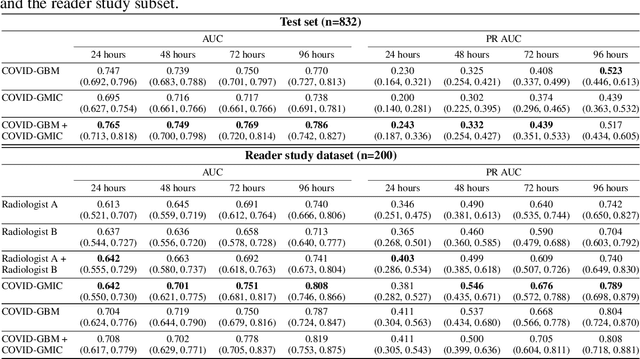
Abstract:During the COVID-19 pandemic, rapid and accurate triage of patients at the emergency department is critical to inform decision-making. We propose a data-driven approach for automatic prediction of deterioration risk using a deep neural network that learns from chest X-ray images, and a gradient boosting model that learns from routine clinical variables. Our AI prognosis system, trained using data from 3,661 patients, achieves an AUC of 0.786 (95% CI: 0.742-0.827) when predicting deterioration within 96 hours. The deep neural network extracts informative areas of chest X-ray images to assist clinicians in interpreting the predictions, and performs comparably to two radiologists in a reader study. In order to verify performance in a real clinical setting, we silently deployed a preliminary version of the deep neural network at NYU Langone Health during the first wave of the pandemic, which produced accurate predictions in real-time. In summary, our findings demonstrate the potential of the proposed system for assisting front-line physicians in the triage of COVID-19 patients.
An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization
Feb 13, 2020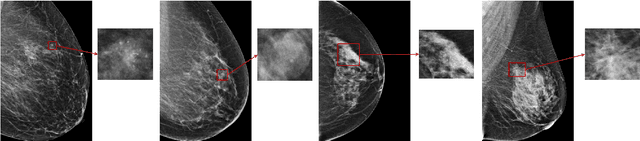
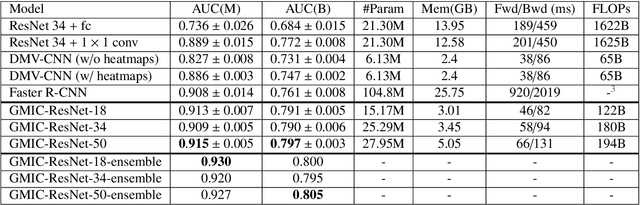
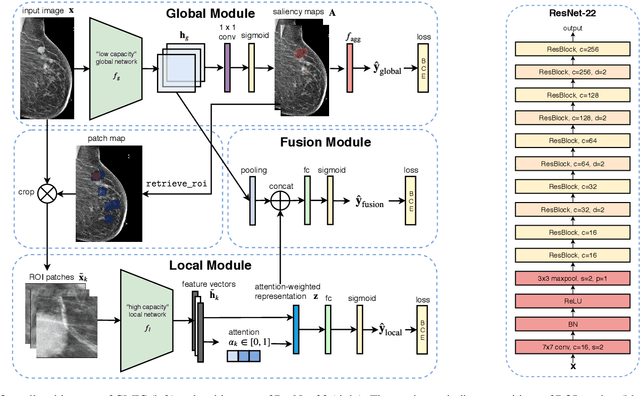
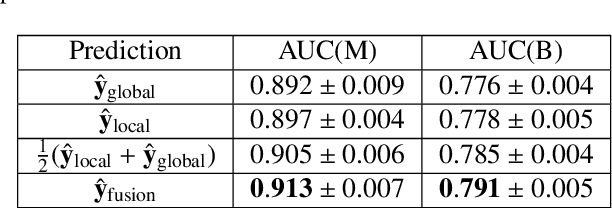
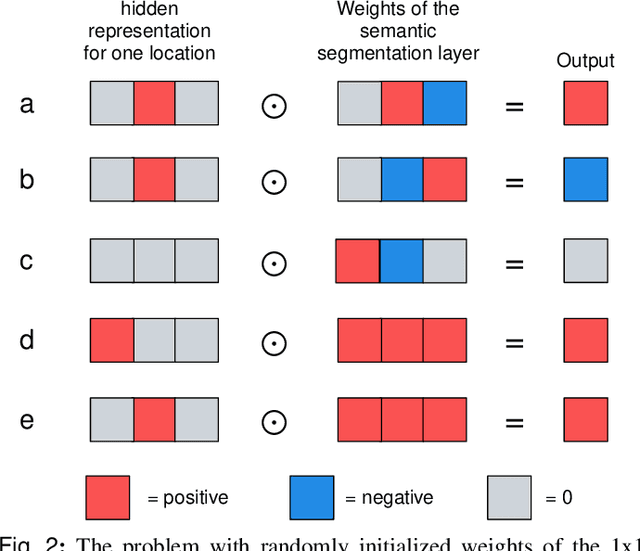
Abstract:Medical images differ from natural images in significantly higher resolutions and smaller regions of interest. Because of these differences, neural network architectures that work well for natural images might not be applicable to medical image analysis. In this work, we extend the globally-aware multiple instance classifier, a framework we proposed to address these unique properties of medical images. This model first uses a low-capacity, yet memory-efficient, network on the whole image to identify the most informative regions. It then applies another higher-capacity network to collect details from chosen regions. Finally, it employs a fusion module that aggregates global and local information to make a final prediction. While existing methods often require lesion segmentation during training, our model is trained with only image-level labels and can generate pixel-level saliency maps indicating possible malignant findings. We apply the model to screening mammography interpretation: predicting the presence or absence of benign and malignant lesions. On the NYU Breast Cancer Screening Dataset, consisting of more than one million images, our model achieves an AUC of 0.93 in classifying breasts with malignant findings, outperforming ResNet-34 and Faster R-CNN. Compared to ResNet-34, our model is 4.1x faster for inference while using 78.4% less GPU memory. Furthermore, we demonstrate, in a reader study, that our model surpasses radiologist-level AUC by a margin of 0.11. The proposed model is available online: https://github.com/nyukat/GMIC.
Screening Mammogram Classification with Prior Exams
Jul 30, 2019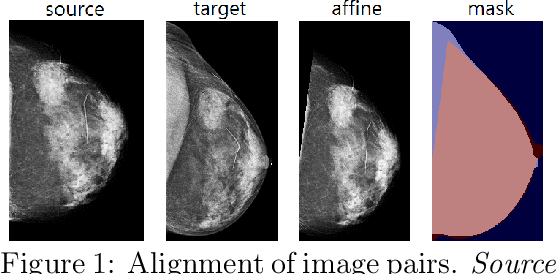
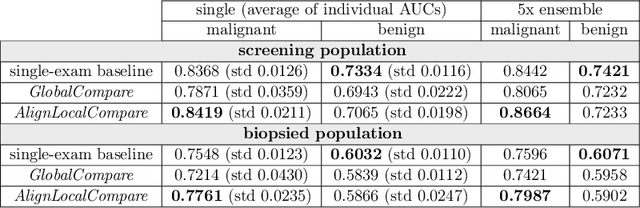
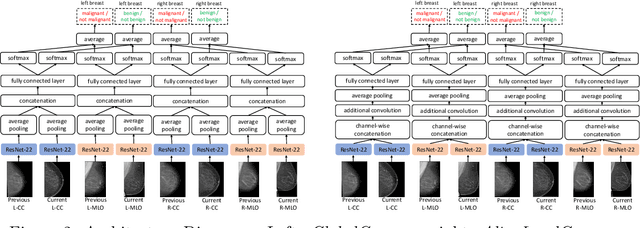
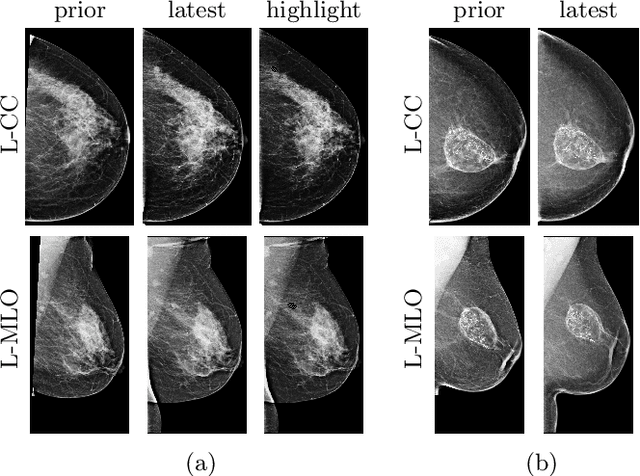
Abstract:Radiologists typically compare a patient's most recent breast cancer screening exam to their previous ones in making informed diagnoses. To reflect this practice, we propose new neural network models that compare pairs of screening mammograms from the same patient. We train and evaluate our proposed models on over 665,000 pairs of images (over 166,000 pairs of exams). Our best model achieves an AUC of 0.866 in predicting malignancy in patients who underwent breast cancer screening, reducing the error rate of the corresponding baseline.
Globally-Aware Multiple Instance Classifier for Breast Cancer Screening
Jun 07, 2019



Abstract:Deep learning models designed for visual classification tasks on natural images have become prevalent in medical image analysis. However, medical images differ from typical natural images in many ways, such as significantly higher resolutions and smaller regions of interest. Moreover, both the global structure and local details play important roles in medical image analysis tasks. To address these unique properties of medical images, we propose a neural network that is able to classify breast cancer lesions utilizing information from both a global saliency map and multiple local patches. The proposed model outperforms the ResNet-based baseline and achieves radiologist-level performance in the interpretation of screening mammography. Although our model is trained only with image-level labels, it is able to generate pixel-level saliency maps that provide localization of possible malignant findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge