Taro Makino
EDINET-Bench: Evaluating LLMs on Complex Financial Tasks using Japanese Financial Statements
Jun 10, 2025



Abstract:Financial analysis presents complex challenges that could leverage large language model (LLM) capabilities. However, the scarcity of challenging financial datasets, particularly for Japanese financial data, impedes academic innovation in financial analytics. As LLMs advance, this lack of accessible research resources increasingly hinders their development and evaluation in this specialized domain. To address this gap, we introduce EDINET-Bench, an open-source Japanese financial benchmark designed to evaluate the performance of LLMs on challenging financial tasks including accounting fraud detection, earnings forecasting, and industry prediction. EDINET-Bench is constructed by downloading annual reports from the past 10 years from Japan's Electronic Disclosure for Investors' NETwork (EDINET) and automatically assigning labels corresponding to each evaluation task. Our experiments reveal that even state-of-the-art LLMs struggle, performing only slightly better than logistic regression in binary classification for fraud detection and earnings forecasting. These results highlight significant challenges in applying LLMs to real-world financial applications and underscore the need for domain-specific adaptation. Our dataset, benchmark construction code, and evaluation code is publicly available to facilitate future research in finance with LLMs.
Supervised Contrastive Block Disentanglement
Feb 11, 2025



Abstract:Real-world datasets often combine data collected under different experimental conditions. This yields larger datasets, but also introduces spurious correlations that make it difficult to model the phenomena of interest. We address this by learning two embeddings to independently represent the phenomena of interest and the spurious correlations. The embedding representing the phenomena of interest is correlated with the target variable $y$, and is invariant to the environment variable $e$. In contrast, the embedding representing the spurious correlations is correlated with $e$. The invariance to $e$ is difficult to achieve on real-world datasets. Our primary contribution is an algorithm called Supervised Contrastive Block Disentanglement (SCBD) that effectively enforces this invariance. It is based purely on Supervised Contrastive Learning, and applies to real-world data better than existing approaches. We empirically validate SCBD on two challenging problems. The first problem is domain generalization, where we achieve strong performance on a synthetic dataset, as well as on Camelyon17-WILDS. We introduce a single hyperparameter $\alpha$ to control the degree of invariance to $e$. When we increase $\alpha$ to strengthen the degree of invariance, out-of-distribution performance improves at the expense of in-distribution performance. The second problem is batch correction, in which we apply SCBD to preserve biological signal and remove inter-well batch effects when modeling single-cell perturbations from 26 million Optical Pooled Screening images.
A Framework for Multi-modal Learning: Jointly Modeling Inter- & Intra-Modality Dependencies
May 27, 2024



Abstract:Supervised multi-modal learning involves mapping multiple modalities to a target label. Previous studies in this field have concentrated on capturing in isolation either the inter-modality dependencies (the relationships between different modalities and the label) or the intra-modality dependencies (the relationships within a single modality and the label). We argue that these conventional approaches that rely solely on either inter- or intra-modality dependencies may not be optimal in general. We view the multi-modal learning problem from the lens of generative models where we consider the target as a source of multiple modalities and the interaction between them. Towards that end, we propose inter- & intra-modality modeling (I2M2) framework, which captures and integrates both the inter- and intra-modality dependencies, leading to more accurate predictions. We evaluate our approach using real-world healthcare and vision-and-language datasets with state-of-the-art models, demonstrating superior performance over traditional methods focusing only on one type of modality dependency.
Generative multitask learning mitigates target-causing confounding
Feb 08, 2022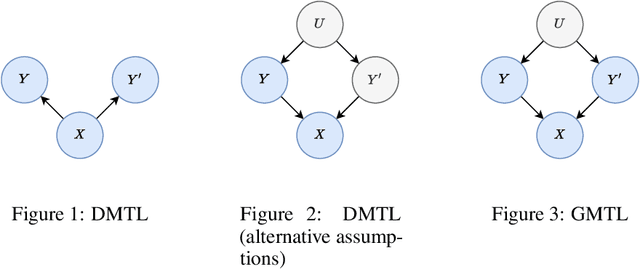


Abstract:We propose a simple and scalable approach to causal representation learning for multitask learning. Our approach requires minimal modification to existing ML systems, and improves robustness to prior probability shift. The improvement comes from mitigating unobserved confounders that cause the targets, but not the input. We refer to them as target-causing confounders. These confounders induce spurious dependencies between the input and targets. This poses a problem for the conventional approach to multitask learning, due to its assumption that the targets are conditionally independent given the input. Our proposed approach takes into account the dependency between the targets in order to alleviate target-causing confounding. All that is required in addition to usual practice is to estimate the joint distribution of the targets to switch from discriminative to generative classification, and to predict all targets jointly. Our results on the Attributes of People and Taskonomy datasets reflect the conceptual improvement in robustness to prior probability shift.
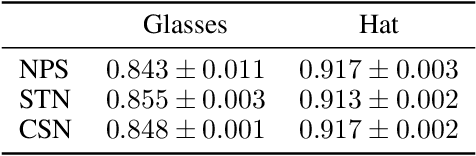
Differences between human and machine perception in medical diagnosis
Nov 28, 2020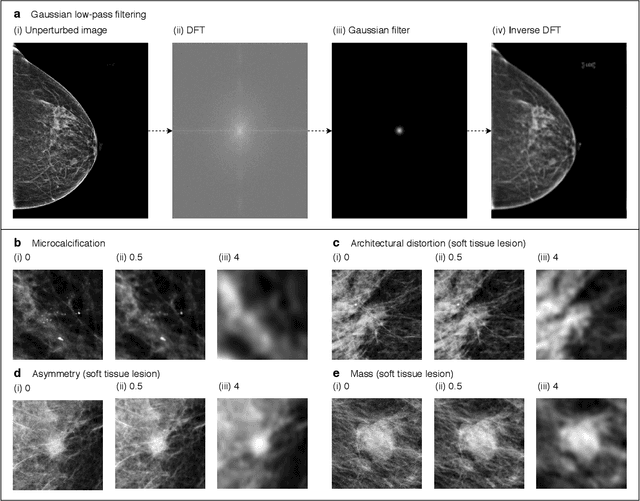
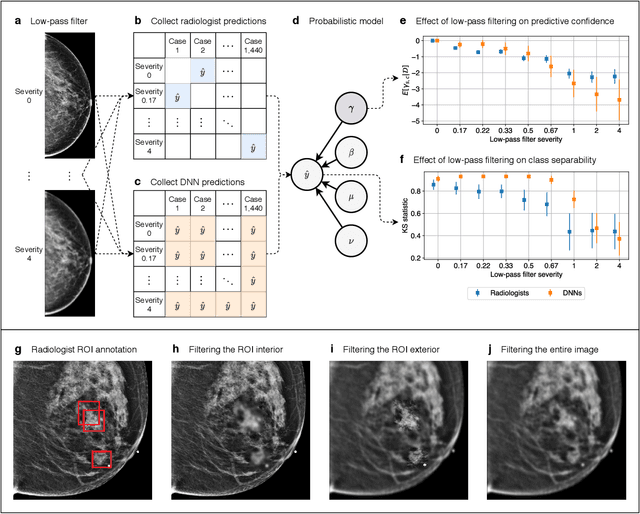
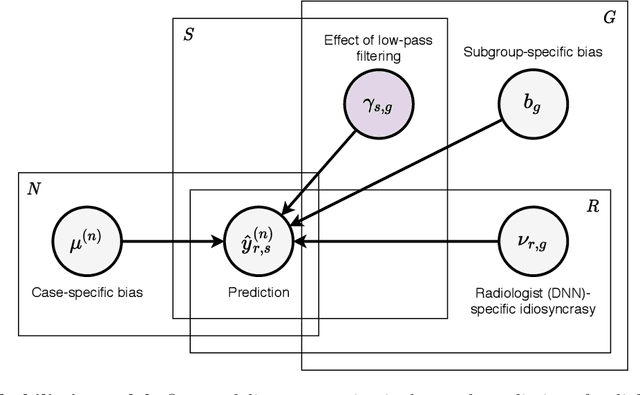
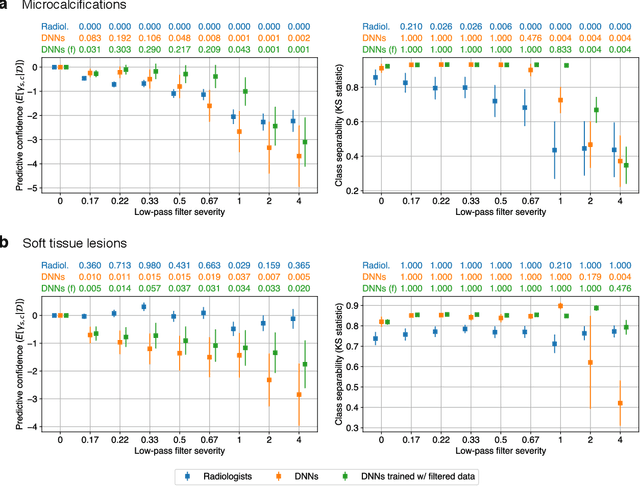
Abstract:Deep neural networks (DNNs) show promise in image-based medical diagnosis, but cannot be fully trusted since their performance can be severely degraded by dataset shifts to which human perception remains invariant. If we can better understand the differences between human and machine perception, we can potentially characterize and mitigate this effect. We therefore propose a framework for comparing human and machine perception in medical diagnosis. The two are compared with respect to their sensitivity to the removal of clinically meaningful information, and to the regions of an image deemed most suspicious. Drawing inspiration from the natural image domain, we frame both comparisons in terms of perturbation robustness. The novelty of our framework is that separate analyses are performed for subgroups with clinically meaningful differences. We argue that this is necessary in order to avert Simpson's paradox and draw correct conclusions. We demonstrate our framework with a case study in breast cancer screening, and reveal significant differences between radiologists and DNNs. We compare the two with respect to their robustness to Gaussian low-pass filtering, performing a subgroup analysis on microcalcifications and soft tissue lesions. For microcalcifications, DNNs use a separate set of high frequency components than radiologists, some of which lie outside the image regions considered most suspicious by radiologists. These features run the risk of being spurious, but if not, could represent potential new biomarkers. For soft tissue lesions, the divergence between radiologists and DNNs is even starker, with DNNs relying heavily on spurious high frequency components ignored by radiologists. Importantly, this deviation in soft tissue lesions was only observable through subgroup analysis, which highlights the importance of incorporating medical domain knowledge into our comparison framework.
Reducing false-positive biopsies with deep neural networks that utilize local and global information in screening mammograms
Sep 19, 2020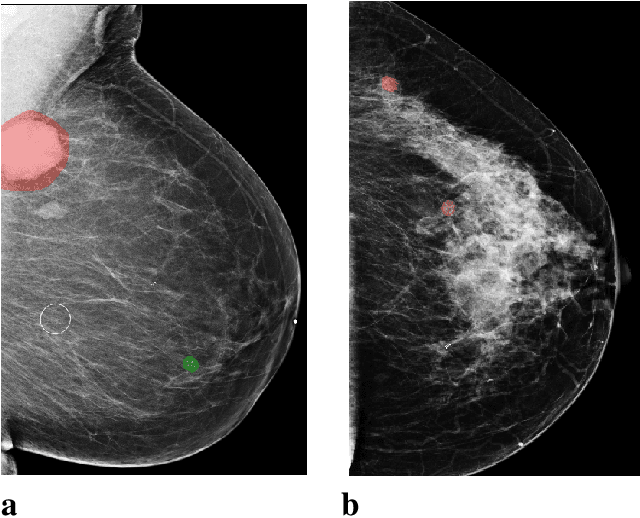
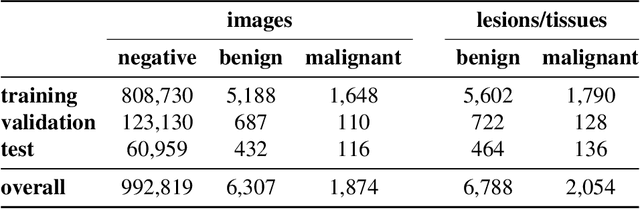
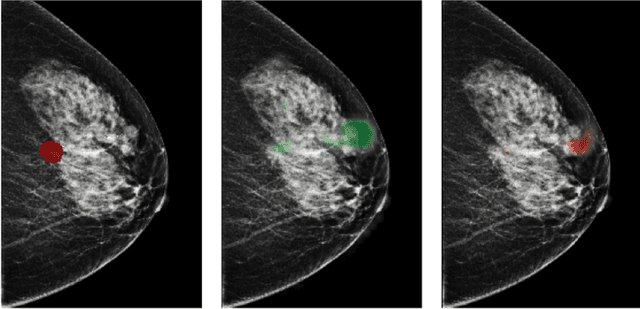
Abstract:Breast cancer is the most common cancer in women, and hundreds of thousands of unnecessary biopsies are done around the world at a tremendous cost. It is crucial to reduce the rate of biopsies that turn out to be benign tissue. In this study, we build deep neural networks (DNNs) to classify biopsied lesions as being either malignant or benign, with the goal of using these networks as second readers serving radiologists to further reduce the number of false positive findings. We enhance the performance of DNNs that are trained to learn from small image patches by integrating global context provided in the form of saliency maps learned from the entire image into their reasoning, similar to how radiologists consider global context when evaluating areas of interest. Our experiments are conducted on a dataset of 229,426 screening mammography exams from 141,473 patients. We achieve an AUC of 0.8 on a test set consisting of 464 benign and 136 malignant lesions.
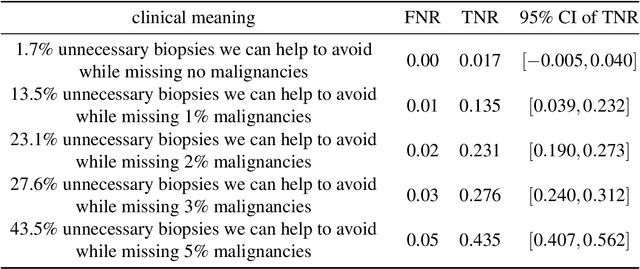
An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department
Aug 04, 2020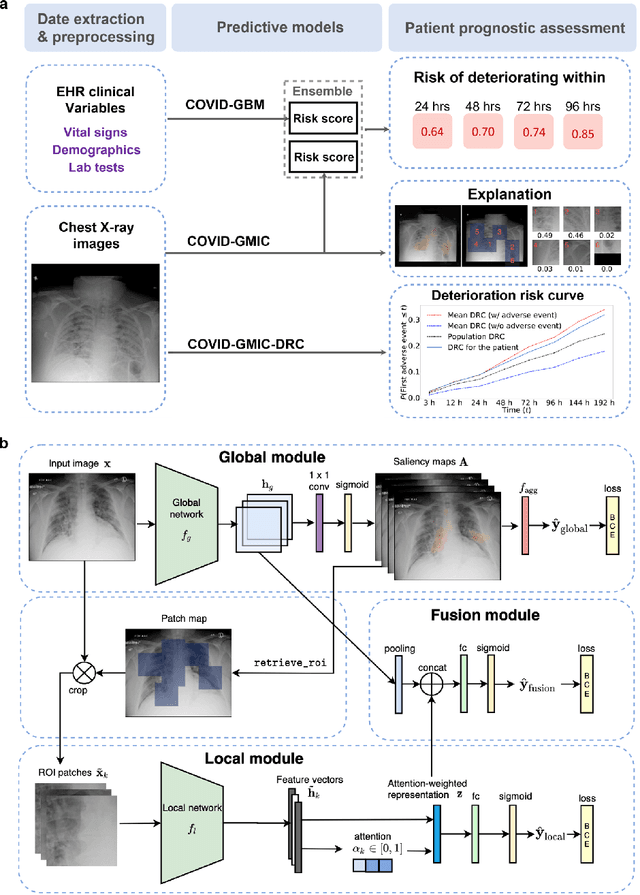
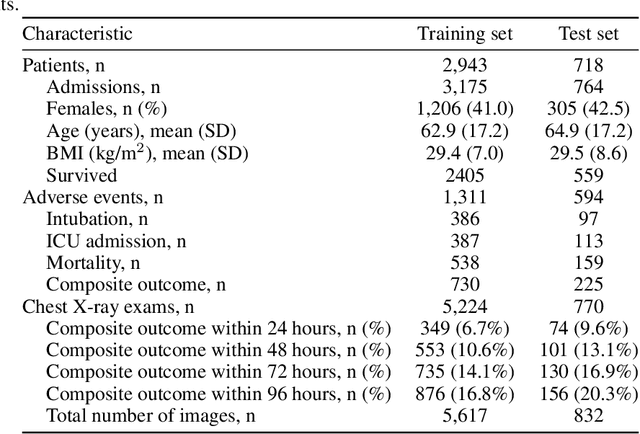
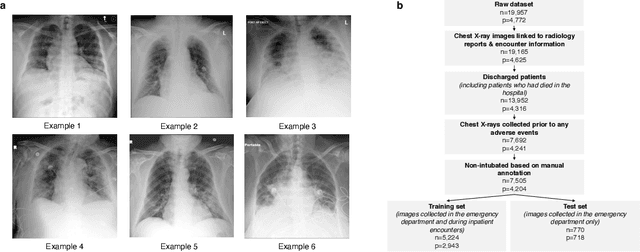
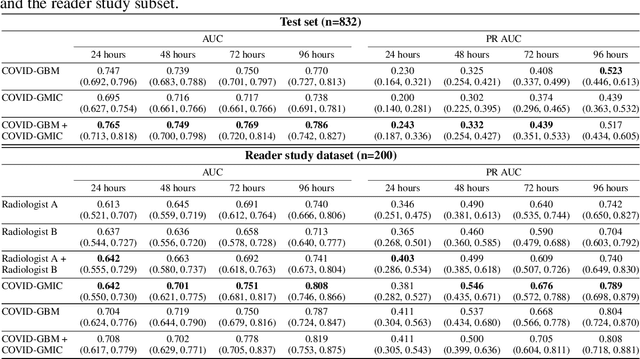
Abstract:During the COVID-19 pandemic, rapid and accurate triage of patients at the emergency department is critical to inform decision-making. We propose a data-driven approach for automatic prediction of deterioration risk using a deep neural network that learns from chest X-ray images, and a gradient boosting model that learns from routine clinical variables. Our AI prognosis system, trained using data from 3,661 patients, achieves an AUC of 0.786 (95% CI: 0.742-0.827) when predicting deterioration within 96 hours. The deep neural network extracts informative areas of chest X-ray images to assist clinicians in interpreting the predictions, and performs comparably to two radiologists in a reader study. In order to verify performance in a real clinical setting, we silently deployed a preliminary version of the deep neural network at NYU Langone Health during the first wave of the pandemic, which produced accurate predictions in real-time. In summary, our findings demonstrate the potential of the proposed system for assisting front-line physicians in the triage of COVID-19 patients.
Understanding the robustness of deep neural network classifiers for breast cancer screening
Mar 23, 2020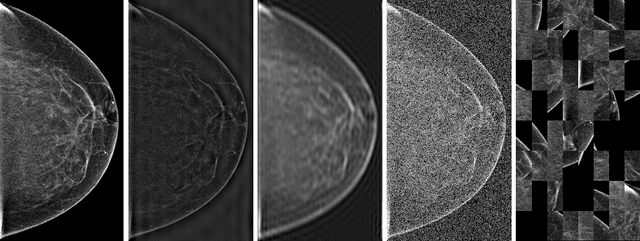
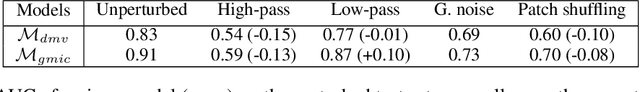
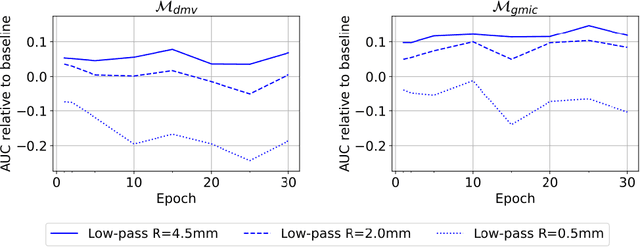
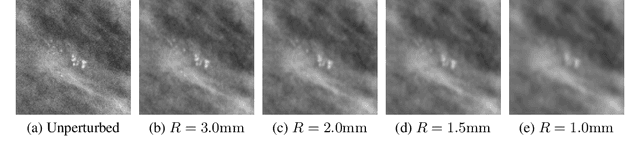
Abstract:Deep neural networks (DNNs) show promise in breast cancer screening, but their robustness to input perturbations must be better understood before they can be clinically implemented. There exists extensive literature on this subject in the context of natural images that can potentially be built upon. However, it cannot be assumed that conclusions about robustness will transfer from natural images to mammogram images, due to significant differences between the two image modalities. In order to determine whether conclusions will transfer, we measure the sensitivity of a radiologist-level screening mammogram image classifier to four commonly studied input perturbations that natural image classifiers are sensitive to. We find that mammogram image classifiers are also sensitive to these perturbations, which suggests that we can build on the existing literature. We also perform a detailed analysis on the effects of low-pass filtering, and find that it degrades the visibility of clinically meaningful features called microcalcifications. Since low-pass filtering removes semantically meaningful information that is predictive of breast cancer, we argue that it is undesirable for mammogram image classifiers to be invariant to it. This is in contrast to natural images, where we do not want DNNs to be sensitive to low-pass filtering due to its tendency to remove information that is human-incomprehensible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge