Heming Yao
Group Contrastive Learning for Weakly Paired Multimodal Data
Feb 03, 2026Abstract:We present GROOVE, a semi-supervised multi-modal representation learning approach for high-content perturbation data where samples across modalities are weakly paired through shared perturbation labels but lack direct correspondence. Our primary contribution is GroupCLIP, a novel group-level contrastive loss that bridges the gap between CLIP for paired cross-modal data and SupCon for uni-modal supervised contrastive learning, addressing a fundamental gap in contrastive learning for weakly-paired settings. We integrate GroupCLIP with an on-the-fly backtranslating autoencoder framework to encourage cross-modally entangled representations while maintaining group-level coherence within a shared latent space. Critically, we introduce a comprehensive combinatorial evaluation framework that systematically assesses representation learners across multiple optimal transport aligners, addressing key limitations in existing evaluation strategies. This framework includes novel simulations that systematically vary shared versus modality-specific perturbation effects enabling principled assessment of method robustness. Our combinatorial benchmarking reveals that there is not yet an aligner that uniformly dominates across settings or modality pairs. Across simulations and two real single-cell genetic perturbation datasets, GROOVE performs on par with or outperforms existing approaches for downstream cross-modal matching and imputation tasks. Our ablation studies demonstrate that GroupCLIP is the key component driving performance gains. These results highlight the importance of leveraging group-level constraints for effective multi-modal representation learning in scenarios where only weak pairing is available.
Supervised Contrastive Block Disentanglement
Feb 11, 2025



Abstract:Real-world datasets often combine data collected under different experimental conditions. This yields larger datasets, but also introduces spurious correlations that make it difficult to model the phenomena of interest. We address this by learning two embeddings to independently represent the phenomena of interest and the spurious correlations. The embedding representing the phenomena of interest is correlated with the target variable $y$, and is invariant to the environment variable $e$. In contrast, the embedding representing the spurious correlations is correlated with $e$. The invariance to $e$ is difficult to achieve on real-world datasets. Our primary contribution is an algorithm called Supervised Contrastive Block Disentanglement (SCBD) that effectively enforces this invariance. It is based purely on Supervised Contrastive Learning, and applies to real-world data better than existing approaches. We empirically validate SCBD on two challenging problems. The first problem is domain generalization, where we achieve strong performance on a synthetic dataset, as well as on Camelyon17-WILDS. We introduce a single hyperparameter $\alpha$ to control the degree of invariance to $e$. When we increase $\alpha$ to strengthen the degree of invariance, out-of-distribution performance improves at the expense of in-distribution performance. The second problem is batch correction, in which we apply SCBD to preserve biological signal and remove inter-well batch effects when modeling single-cell perturbations from 26 million Optical Pooled Screening images.
Gene-Level Representation Learning via Interventional Style Transfer in Optical Pooled Screening
Jun 11, 2024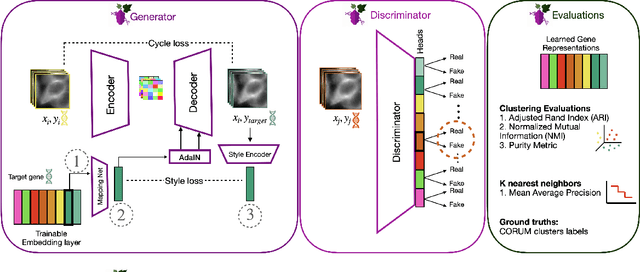
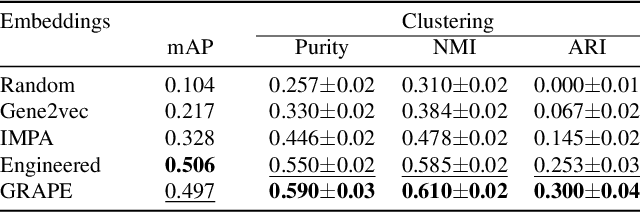
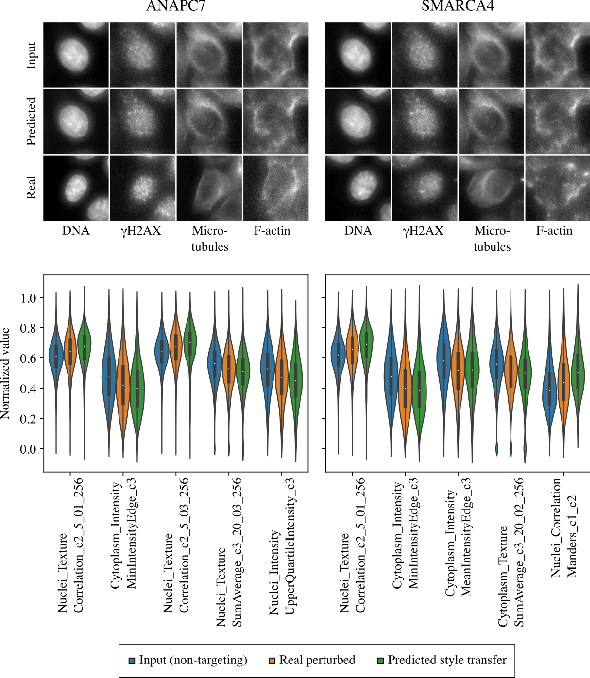

Abstract:Optical pooled screening (OPS) combines automated microscopy and genetic perturbations to systematically study gene function in a scalable and cost-effective way. Leveraging the resulting data requires extracting biologically informative representations of cellular perturbation phenotypes from images. We employ a style-transfer approach to learn gene-level feature representations from images of genetically perturbed cells obtained via OPS. Our method outperforms widely used engineered features in clustering gene representations according to gene function, demonstrating its utility for uncovering latent biological relationships. This approach offers a promising alternative to investigate the role of genes in health and disease.
Weakly Supervised Set-Consistency Learning Improves Morphological Profiling of Single-Cell Images
Jun 08, 2024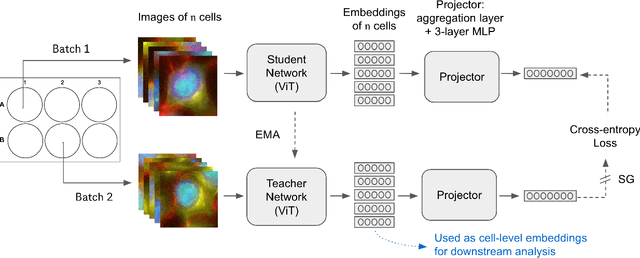



Abstract:Optical Pooled Screening (OPS) is a powerful tool combining high-content microscopy with genetic engineering to investigate gene function in disease. The characterization of high-content images remains an active area of research and is currently undergoing rapid innovation through the application of self-supervised learning and vision transformers. In this study, we propose a set-level consistency learning algorithm, Set-DINO, that combines self-supervised learning with weak supervision to improve learned representations of perturbation effects in single-cell images. Our method leverages the replicate structure of OPS experiments (i.e., cells undergoing the same genetic perturbation, both within and across batches) as a form of weak supervision. We conduct extensive experiments on a large-scale OPS dataset with more than 5000 genetic perturbations, and demonstrate that Set-DINO helps mitigate the impact of confounders and encodes more biologically meaningful information. In particular, Set-DINO recalls known biological relationships with higher accuracy compared to commonly used methods for morphological profiling, suggesting that it can generate more reliable insights from drug target discovery campaigns leveraging OPS.
Unsupervised Segmentation of Colonoscopy Images
Dec 19, 2023



Abstract:Colonoscopy plays a crucial role in the diagnosis and prognosis of various gastrointestinal diseases. Due to the challenges of collecting large-scale high-quality ground truth annotations for colonoscopy images, and more generally medical images, we explore using self-supervised features from vision transformers in three challenging tasks for colonoscopy images. Our results indicate that image-level features learned from DINO models achieve image classification performance comparable to fully supervised models, and patch-level features contain rich semantic information for object detection. Furthermore, we demonstrate that self-supervised features combined with unsupervised segmentation can be used to discover multiple clinically relevant structures in a fully unsupervised manner, demonstrating the tremendous potential of applying these methods in medical image analysis.
A Novel Tropical Geometry-based Interpretable Machine Learning Method: Application in Prognosis of Advanced Heart Failure
Dec 09, 2021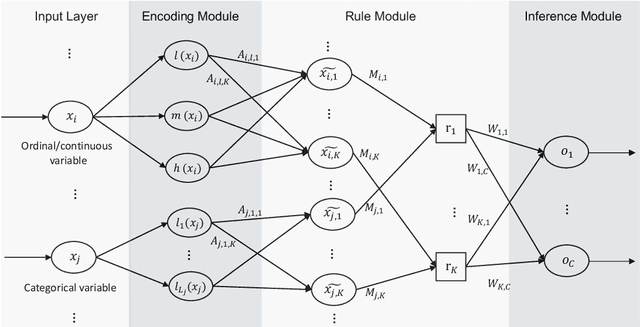
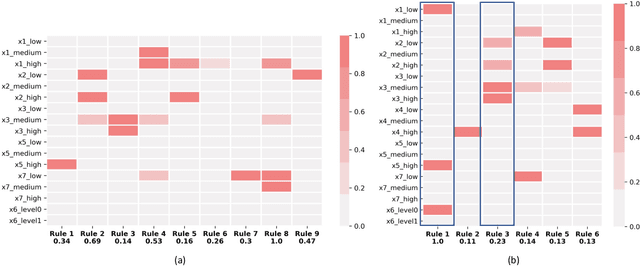
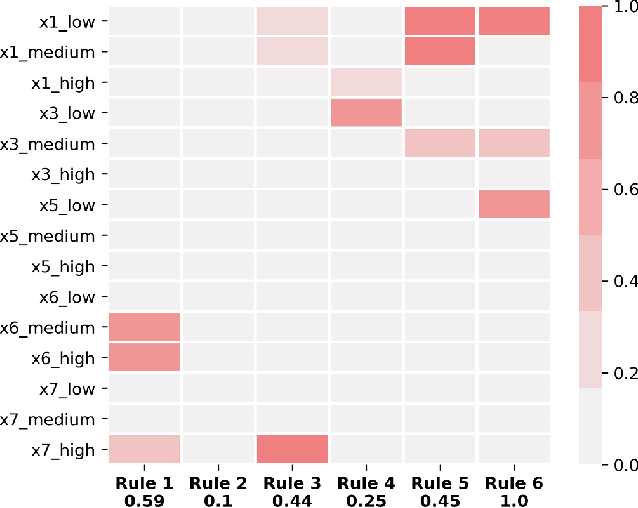
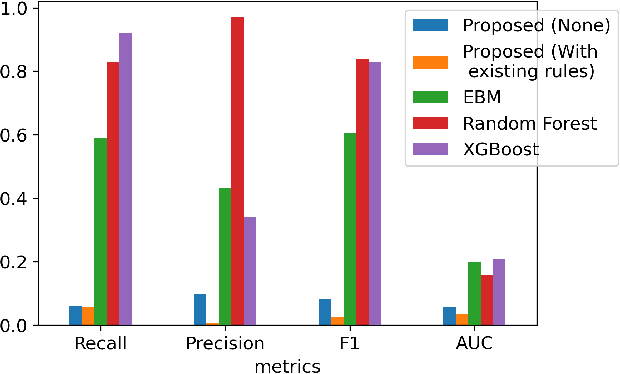
Abstract:A model's interpretability is essential to many practical applications such as clinical decision support systems. In this paper, a novel interpretable machine learning method is presented, which can model the relationship between input variables and responses in humanly understandable rules. The method is built by applying tropical geometry to fuzzy inference systems, wherein variable encoding functions and salient rules can be discovered by supervised learning. Experiments using synthetic datasets were conducted to investigate the performance and capacity of the proposed algorithm in classification and rule discovery. Furthermore, the proposed method was applied to a clinical application that identified heart failure patients that would benefit from advanced therapies such as heart transplant or durable mechanical circulatory support. Experimental results show that the proposed network achieved great performance on the classification tasks. In addition to learning humanly understandable rules from the dataset, existing fuzzy domain knowledge can be easily transferred into the network and used to facilitate model training. From our results, the proposed model and the ability of learning existing domain knowledge can significantly improve the model generalizability. The characteristics of the proposed network make it promising in applications requiring model reliability and justification.
Motion-based Camera Localization System in Colonoscopy Videos
Dec 04, 2020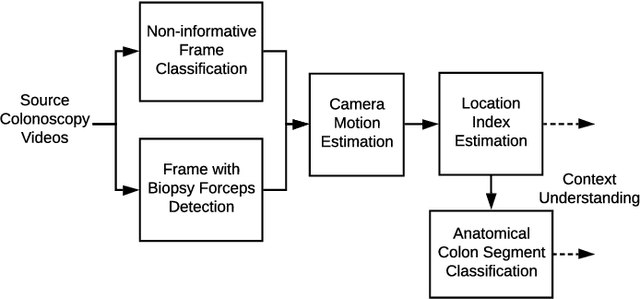
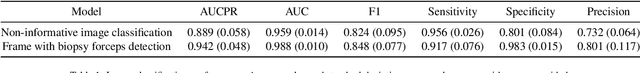
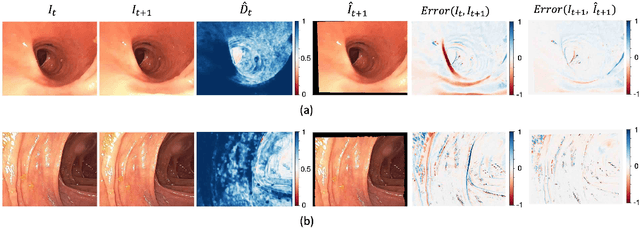
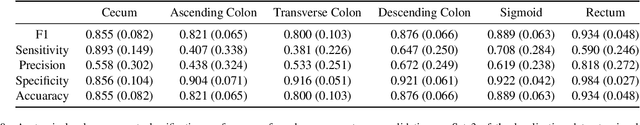
Abstract:Optical colonoscopy is an essential diagnostic and prognostic tool for many gastrointestinal diseases including cancer screening and staging, intestinal bleeding, diarrhea, abdominal symptom evaluation, and inflammatory bowel disease assessment. However, the evaluation, classification, and quantification of findings on colonoscopy are subject to inter-observer variation. Automated assessment of colonoscopy is of interest considering the subjectivity present in qualitative human interpretations of colonoscopy findings. Localization of the camera is an essential element to consider when inferring the meaning and context of findings for diseases evaluated by colonoscopy. In this study, we proposed a camera localization system to estimate the approximate anatomic location of the camera and classify the anatomical colon segment the camera is in. The camera localization system starts with non-informative frame detection to remove frames without camera motion information. Then a self-training end-to-end convolutional neural network was built to estimate the camera motion. With the estimated camera motion, the camera trajectory can be derived, and the location index can be calculated. Based on the estimated location index, anatomical colon segment classification was performed by building the colon template. The algorithm was trained and validated using colonoscopy videos collected from routine clinical practice. From our results, the average accuracy of the classification is 0.759, which is substantially higher than the performance of using the location index built from other methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge