David Richmond
Contextualizing biological perturbation experiments through language
Feb 28, 2025
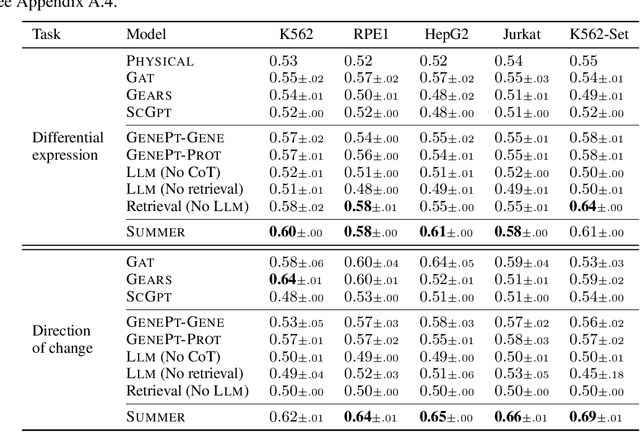

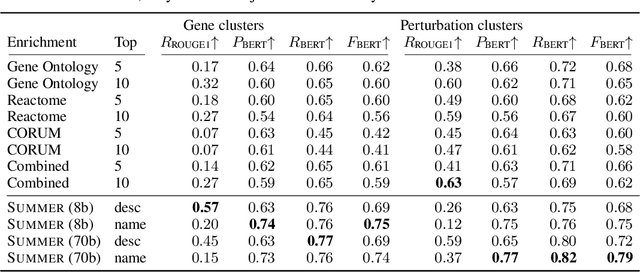
Abstract:High-content perturbation experiments allow scientists to probe biomolecular systems at unprecedented resolution, but experimental and analysis costs pose significant barriers to widespread adoption. Machine learning has the potential to guide efficient exploration of the perturbation space and extract novel insights from these data. However, current approaches neglect the semantic richness of the relevant biology, and their objectives are misaligned with downstream biological analyses. In this paper, we hypothesize that large language models (LLMs) present a natural medium for representing complex biological relationships and rationalizing experimental outcomes. We propose PerturbQA, a benchmark for structured reasoning over perturbation experiments. Unlike current benchmarks that primarily interrogate existing knowledge, PerturbQA is inspired by open problems in perturbation modeling: prediction of differential expression and change of direction for unseen perturbations, and gene set enrichment. We evaluate state-of-the-art machine learning and statistical approaches for modeling perturbations, as well as standard LLM reasoning strategies, and we find that current methods perform poorly on PerturbQA. As a proof of feasibility, we introduce Summer (SUMMarize, retrievE, and answeR, a simple, domain-informed LLM framework that matches or exceeds the current state-of-the-art. Our code and data are publicly available at https://github.com/genentech/PerturbQA.
Supervised Contrastive Block Disentanglement
Feb 11, 2025



Abstract:Real-world datasets often combine data collected under different experimental conditions. This yields larger datasets, but also introduces spurious correlations that make it difficult to model the phenomena of interest. We address this by learning two embeddings to independently represent the phenomena of interest and the spurious correlations. The embedding representing the phenomena of interest is correlated with the target variable $y$, and is invariant to the environment variable $e$. In contrast, the embedding representing the spurious correlations is correlated with $e$. The invariance to $e$ is difficult to achieve on real-world datasets. Our primary contribution is an algorithm called Supervised Contrastive Block Disentanglement (SCBD) that effectively enforces this invariance. It is based purely on Supervised Contrastive Learning, and applies to real-world data better than existing approaches. We empirically validate SCBD on two challenging problems. The first problem is domain generalization, where we achieve strong performance on a synthetic dataset, as well as on Camelyon17-WILDS. We introduce a single hyperparameter $\alpha$ to control the degree of invariance to $e$. When we increase $\alpha$ to strengthen the degree of invariance, out-of-distribution performance improves at the expense of in-distribution performance. The second problem is batch correction, in which we apply SCBD to preserve biological signal and remove inter-well batch effects when modeling single-cell perturbations from 26 million Optical Pooled Screening images.
Learning Identifiable Factorized Causal Representations of Cellular Responses
Oct 29, 2024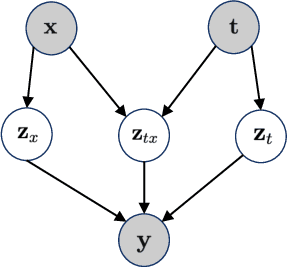
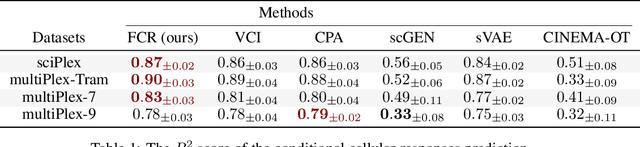
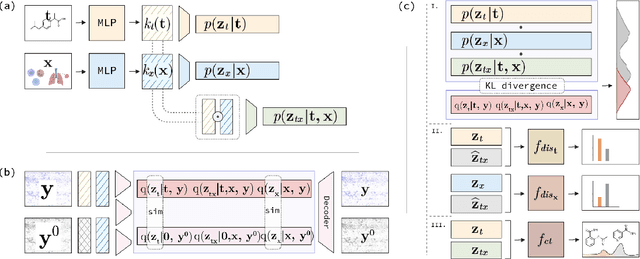

Abstract:The study of cells and their responses to genetic or chemical perturbations promises to accelerate the discovery of therapeutic targets. However, designing adequate and insightful models for such data is difficult because the response of a cell to perturbations essentially depends on its biological context (e.g., genetic background or cell type). For example, while discovering therapeutic targets, one may want to enrich for drugs that specifically target a certain cell type. This challenge emphasizes the need for methods that explicitly take into account potential interactions between drugs and contexts. Towards this goal, we propose a novel Factorized Causal Representation (FCR) learning method that reveals causal structure in single-cell perturbation data from several cell lines. Based on the framework of identifiable deep generative models, FCR learns multiple cellular representations that are disentangled, comprised of covariate-specific ($\mathbf{z}_x$), treatment-specific ($\mathbf{z}_{t}$), and interaction-specific ($\mathbf{z}_{tx}$) blocks. Based on recent advances in non-linear ICA theory, we prove the component-wise identifiability of $\mathbf{z}_{tx}$ and block-wise identifiability of $\mathbf{z}_t$ and $\mathbf{z}_x$. Then, we present our implementation of FCR, and empirically demonstrate that it outperforms state-of-the-art baselines in various tasks across four single-cell datasets.
Gene-Level Representation Learning via Interventional Style Transfer in Optical Pooled Screening
Jun 11, 2024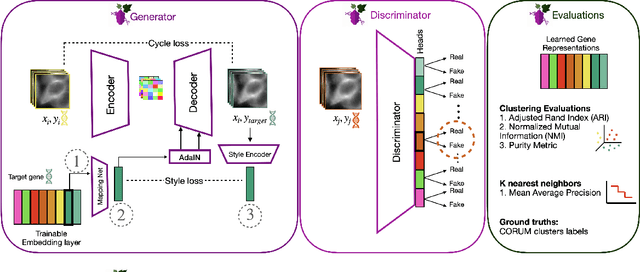
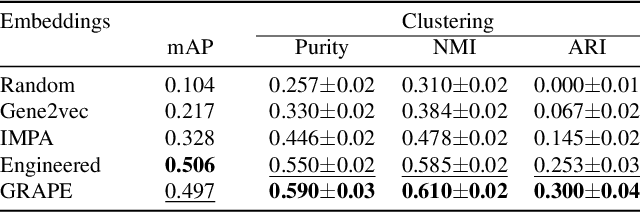
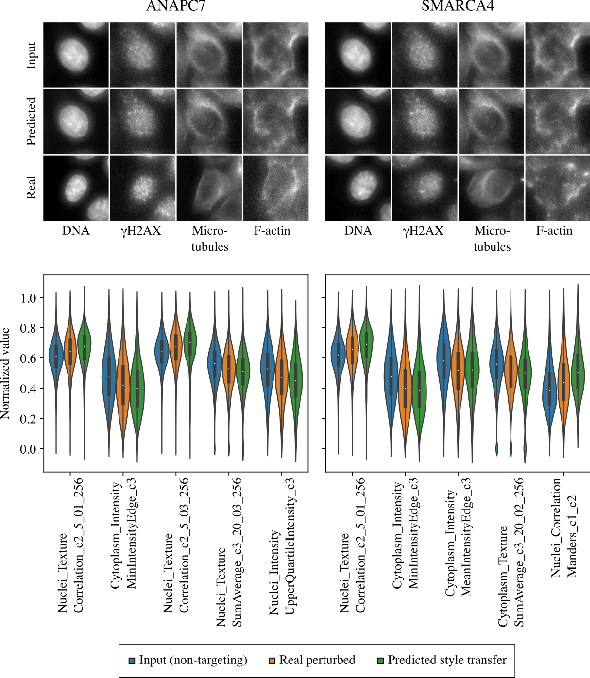

Abstract:Optical pooled screening (OPS) combines automated microscopy and genetic perturbations to systematically study gene function in a scalable and cost-effective way. Leveraging the resulting data requires extracting biologically informative representations of cellular perturbation phenotypes from images. We employ a style-transfer approach to learn gene-level feature representations from images of genetically perturbed cells obtained via OPS. Our method outperforms widely used engineered features in clustering gene representations according to gene function, demonstrating its utility for uncovering latent biological relationships. This approach offers a promising alternative to investigate the role of genes in health and disease.
Weakly Supervised Set-Consistency Learning Improves Morphological Profiling of Single-Cell Images
Jun 08, 2024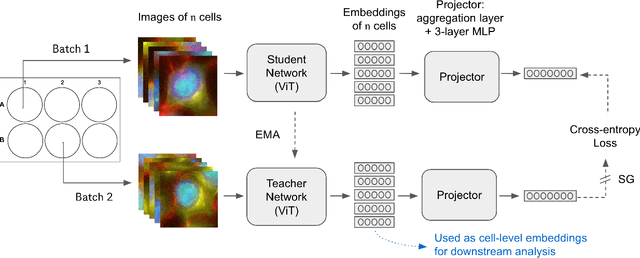



Abstract:Optical Pooled Screening (OPS) is a powerful tool combining high-content microscopy with genetic engineering to investigate gene function in disease. The characterization of high-content images remains an active area of research and is currently undergoing rapid innovation through the application of self-supervised learning and vision transformers. In this study, we propose a set-level consistency learning algorithm, Set-DINO, that combines self-supervised learning with weak supervision to improve learned representations of perturbation effects in single-cell images. Our method leverages the replicate structure of OPS experiments (i.e., cells undergoing the same genetic perturbation, both within and across batches) as a form of weak supervision. We conduct extensive experiments on a large-scale OPS dataset with more than 5000 genetic perturbations, and demonstrate that Set-DINO helps mitigate the impact of confounders and encodes more biologically meaningful information. In particular, Set-DINO recalls known biological relationships with higher accuracy compared to commonly used methods for morphological profiling, suggesting that it can generate more reliable insights from drug target discovery campaigns leveraging OPS.
Unsupervised Segmentation of Colonoscopy Images
Dec 19, 2023



Abstract:Colonoscopy plays a crucial role in the diagnosis and prognosis of various gastrointestinal diseases. Due to the challenges of collecting large-scale high-quality ground truth annotations for colonoscopy images, and more generally medical images, we explore using self-supervised features from vision transformers in three challenging tasks for colonoscopy images. Our results indicate that image-level features learned from DINO models achieve image classification performance comparable to fully supervised models, and patch-level features contain rich semantic information for object detection. Furthermore, we demonstrate that self-supervised features combined with unsupervised segmentation can be used to discover multiple clinically relevant structures in a fully unsupervised manner, demonstrating the tremendous potential of applying these methods in medical image analysis.
Deep Learning based detection of Acute Aortic Syndrome in contrast CT images
Apr 03, 2020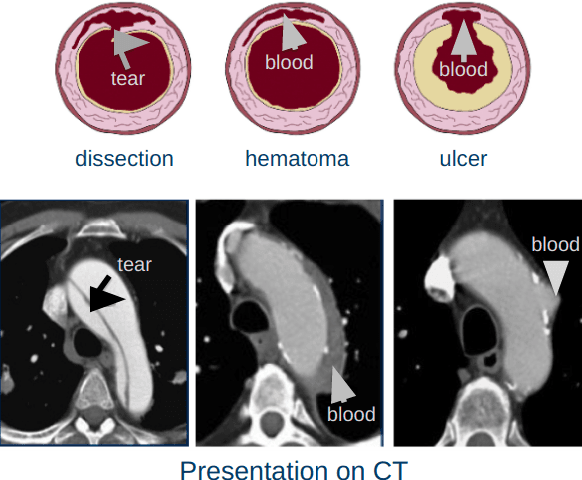
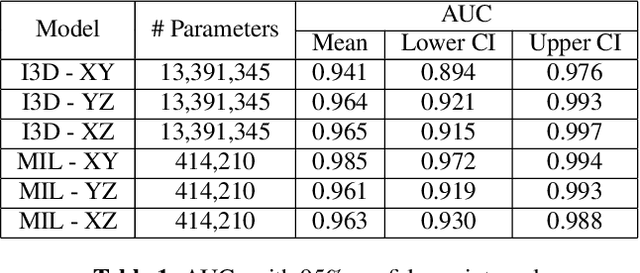
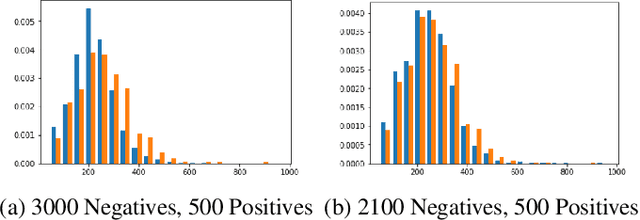
Abstract:Acute aortic syndrome (AAS) is a group of life threatening conditions of the aorta. We have developed an end-to-end automatic approach to detect AAS in computed tomography (CT) images. Our approach consists of two steps. At first, we extract N cross sections along the segmented aorta centerline for each CT scan. These cross sections are stacked together to form a new volume which is then classified using two different classifiers, a 3D convolutional neural network (3D CNN) and a multiple instance learning (MIL). We trained, validated, and compared two models on 2291 contrast CT volumes. We tested on a set aside cohort of 230 normal and 50 positive CT volumes. Our models detected AAS with an Area under Receiver Operating Characteristic curve (AUC) of 0.965 and 0.985 using 3DCNN and MIL, respectively.
DeadNet: Identifying Phototoxicity from Label-free Microscopy Images of Cells using Deep ConvNets
Jan 22, 2017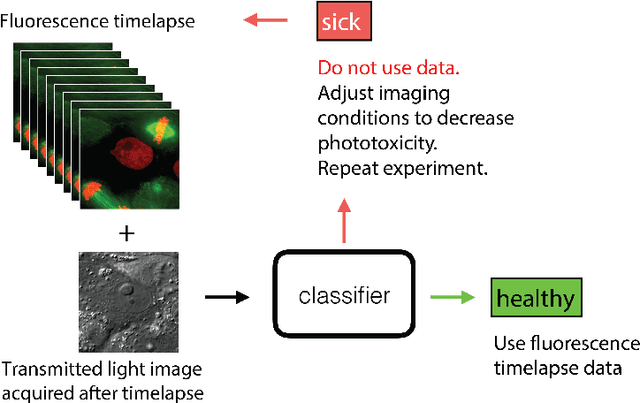
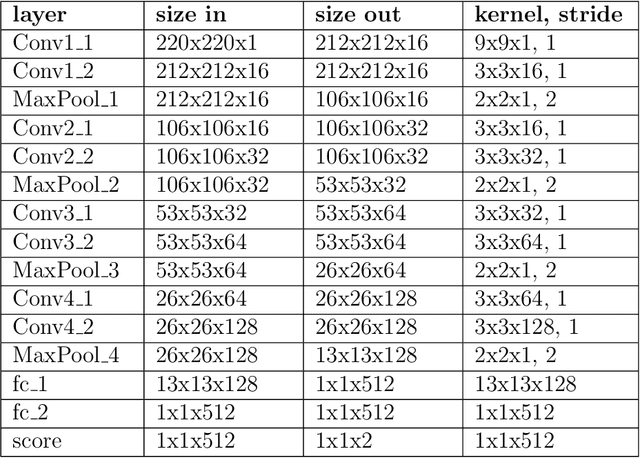
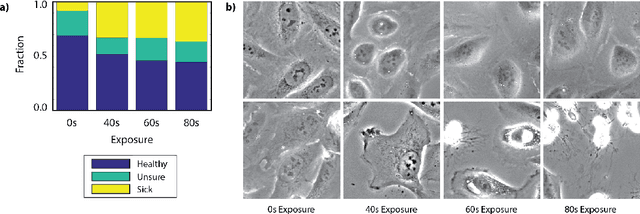
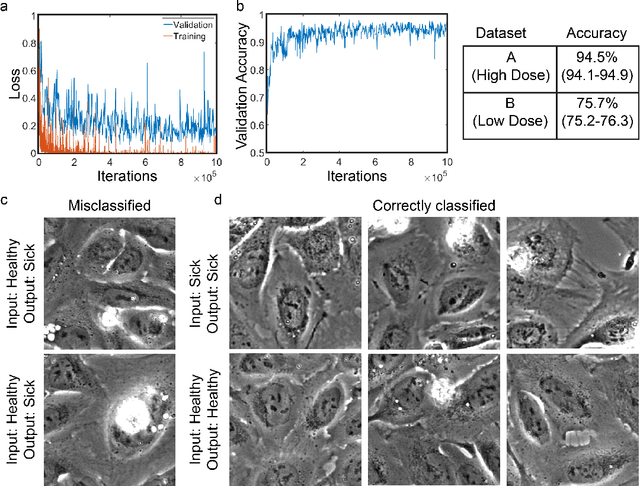
Abstract:Exposure to intense illumination light is an unavoidable consequence of fluorescence microscopy, and poses a risk to the health of the sample in every live-cell fluorescence microscopy experiment. Furthermore, the possible side-effects of phototoxicity on the scientific conclusions that are drawn from an imaging experiment are often unaccounted for. Previously, controlling for phototoxicity in imaging experiments required additional labels and experiments, limiting its widespread application. Here we provide a proof-of-principle demonstration that the phototoxic effects of an imaging experiment can be identified directly from a single phase-contrast image using deep convolutional neural networks (ConvNets). This lays the groundwork for an automated tool for assessing cell health in a wide range of imaging experiments. Interpretability of such a method is crucial for its adoption. We take steps towards interpreting the classification mechanism of the trained ConvNet by visualizing salient features of images that contribute to accurate classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge