Menghua Wu
Advertising in AI systems: Society must be vigilant
May 23, 2025Abstract:AI systems have increasingly become our gateways to the Internet. We argue that just as advertising has driven the monetization of web search and social media, so too will commercial incentives shape the content served by AI. Unlike traditional media, however, the outputs of these systems are dynamic, personalized, and lack clear provenance -- raising concerns for transparency and regulation. In this paper, we envision how commercial content could be delivered through generative AI-based systems. Based on the requirements of key stakeholders -- advertisers, consumers, and platforms -- we propose design principles for commercially-influenced AI systems. We then outline high-level strategies for end users to identify and mitigate commercial biases from model outputs. Finally, we conclude with open questions and a call to action towards these goals.
Thought calibration: Efficient and confident test-time scaling
May 23, 2025Abstract:Reasoning large language models achieve impressive test-time scaling by thinking for longer, but this performance gain comes at significant compute cost. Directly limiting test-time budget hurts overall performance, but not all problems are equally difficult. We propose thought calibration to decide dynamically when thinking can be terminated. To calibrate our decision rule, we view a language model's growing body of thoughts as a nested sequence of reasoning trees, where the goal is to identify the point at which novel reasoning plateaus. We realize this framework through lightweight probes that operate on top of the language model's hidden representations, which are informative of both the reasoning structure and overall consistency of response. Based on three reasoning language models and four datasets, thought calibration preserves model performance with up to a 60% reduction in thinking tokens on in-distribution data, and up to 20% in out-of-distribution data.
Contextualizing biological perturbation experiments through language
Feb 28, 2025
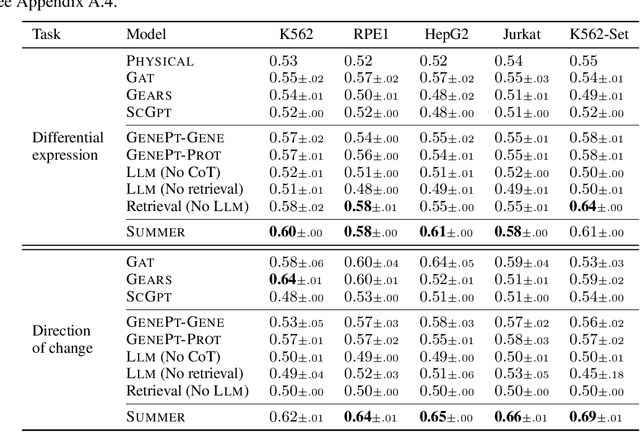

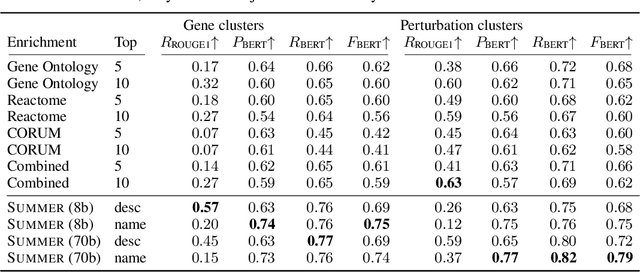
Abstract:High-content perturbation experiments allow scientists to probe biomolecular systems at unprecedented resolution, but experimental and analysis costs pose significant barriers to widespread adoption. Machine learning has the potential to guide efficient exploration of the perturbation space and extract novel insights from these data. However, current approaches neglect the semantic richness of the relevant biology, and their objectives are misaligned with downstream biological analyses. In this paper, we hypothesize that large language models (LLMs) present a natural medium for representing complex biological relationships and rationalizing experimental outcomes. We propose PerturbQA, a benchmark for structured reasoning over perturbation experiments. Unlike current benchmarks that primarily interrogate existing knowledge, PerturbQA is inspired by open problems in perturbation modeling: prediction of differential expression and change of direction for unseen perturbations, and gene set enrichment. We evaluate state-of-the-art machine learning and statistical approaches for modeling perturbations, as well as standard LLM reasoning strategies, and we find that current methods perform poorly on PerturbQA. As a proof of feasibility, we introduce Summer (SUMMarize, retrievE, and answeR, a simple, domain-informed LLM framework that matches or exceeds the current state-of-the-art. Our code and data are publicly available at https://github.com/genentech/PerturbQA.
Predicting sub-population specific viral evolution
Oct 28, 2024



Abstract:Forecasting the change in the distribution of viral variants is crucial for therapeutic design and disease surveillance. This task poses significant modeling challenges due to the sharp differences in virus distributions across sub-populations (e.g., countries) and their dynamic interactions. Existing machine learning approaches that model the variant distribution as a whole are incapable of making location-specific predictions and ignore transmissions that shape the viral landscape. In this paper, we propose a sub-population specific protein evolution model, which predicts the time-resolved distributions of viral proteins in different locations. The algorithm explicitly models the transmission rates between sub-populations and learns their interdependence from data. The change in protein distributions across all sub-populations is defined through a linear ordinary differential equation (ODE) parametrized by transmission rates. Solving this ODE yields the likelihood of a given protein occurring in particular sub-populations. Multi-year evaluation on both SARS-CoV-2 and influenza A/H3N2 demonstrates that our model outperforms baselines in accurately predicting distributions of viral proteins across continents and countries. We also find that the transmission rates learned from data are consistent with the transmission pathways discovered by retrospective phylogenetic analysis.
Learning to refine domain knowledge for biological network inference
Oct 18, 2024



Abstract:Perturbation experiments allow biologists to discover causal relationships between variables of interest, but the sparsity and high dimensionality of these data pose significant challenges for causal structure learning algorithms. Biological knowledge graphs can bootstrap the inference of causal structures in these situations, but since they compile vastly diverse information, they can bias predictions towards well-studied systems. Alternatively, amortized causal structure learning algorithms encode inductive biases through data simulation and train supervised models to recapitulate these synthetic graphs. However, realistically simulating biology is arguably even harder than understanding a specific system. In this work, we take inspiration from both strategies and propose an amortized algorithm for refining domain knowledge, based on data observations. On real and synthetic datasets, we show that our approach outperforms baselines in recovering ground truth causal graphs and identifying errors in the prior knowledge with limited interventional data.
Predicting perturbation targets with causal differential networks
Oct 04, 2024



Abstract:Rationally identifying variables responsible for changes to a biological system can enable myriad applications in disease understanding and cell engineering. From a causality perspective, we are given two datasets generated by the same causal model, one observational (control) and one interventional (perturbed). The goal is to isolate the subset of measured variables (e.g. genes) that were the targets of the intervention, i.e. those whose conditional independencies have changed. Knowing the causal graph would limit the search space, allowing us to efficiently pinpoint these variables. However, current algorithms that infer causal graphs in the presence of unknown intervention targets scale poorly to the hundreds or thousands of variables in biological data, as they must jointly search the combinatorial spaces of graphs and consistent intervention targets. In this work, we propose a causality-inspired approach for predicting perturbation targets that decouples the two search steps. First, we use an amortized causal discovery model to separately infer causal graphs from the observational and interventional datasets. Then, we learn to map these paired graphs to the sets of variables that were intervened upon, in a supervised learning framework. This approach consistently outperforms baselines for perturbation modeling on seven single-cell transcriptomics datasets, each with thousands of measured variables. We also demonstrate significant improvements over six causal discovery algorithms in predicting intervention targets across a variety of tractable, synthetic datasets.
Sample, estimate, aggregate: A recipe for causal discovery foundation models
Feb 02, 2024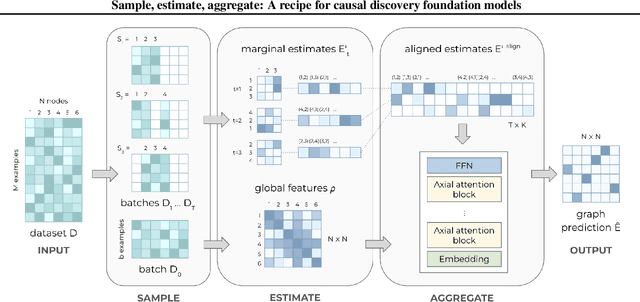
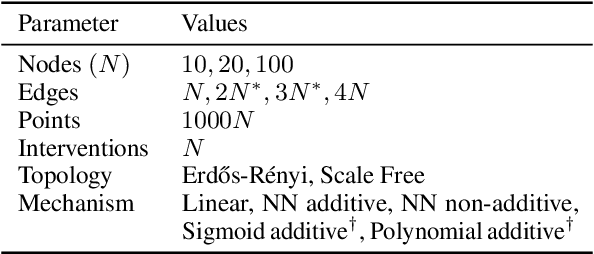
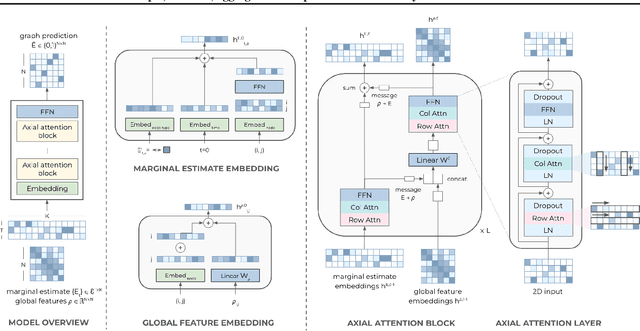
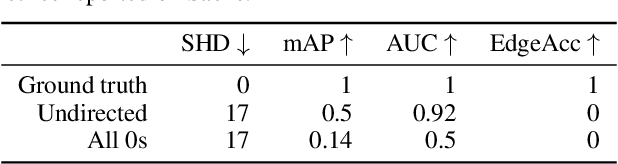
Abstract:Causal discovery, the task of inferring causal structure from data, promises to accelerate scientific research, inform policy making, and more. However, the per-dataset nature of existing causal discovery algorithms renders them slow, data hungry, and brittle. Inspired by foundation models, we propose a causal discovery framework where a deep learning model is pretrained to resolve predictions from classical discovery algorithms run over smaller subsets of variables. This method is enabled by the observations that the outputs from classical algorithms are fast to compute for small problems, informative of (marginal) data structure, and their structure outputs as objects remain comparable across datasets. Our method achieves state-of-the-art performance on synthetic and realistic datasets, generalizes to data generating mechanisms not seen during training, and offers inference speeds that are orders of magnitude faster than existing models.
High-Fidelity 3D Face Generation from Natural Language Descriptions
May 05, 2023
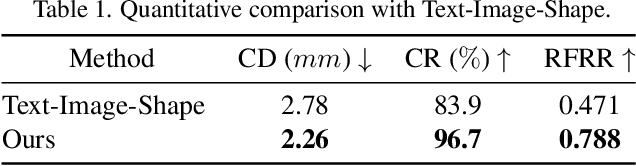
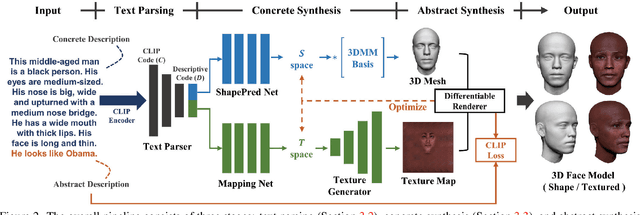
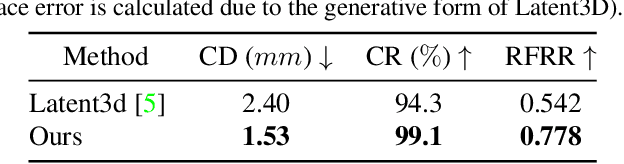
Abstract:Synthesizing high-quality 3D face models from natural language descriptions is very valuable for many applications, including avatar creation, virtual reality, and telepresence. However, little research ever tapped into this task. We argue the major obstacle lies in 1) the lack of high-quality 3D face data with descriptive text annotation, and 2) the complex mapping relationship between descriptive language space and shape/appearance space. To solve these problems, we build Describe3D dataset, the first large-scale dataset with fine-grained text descriptions for text-to-3D face generation task. Then we propose a two-stage framework to first generate a 3D face that matches the concrete descriptions, then optimize the parameters in the 3D shape and texture space with abstract description to refine the 3D face model. Extensive experimental results show that our method can produce a faithful 3D face that conforms to the input descriptions with higher accuracy and quality than previous methods. The code and Describe3D dataset are released at https://github.com/zhuhao-nju/describe3d .
DiffDock-PP: Rigid Protein-Protein Docking with Diffusion Models
Apr 08, 2023



Abstract:Understanding how proteins structurally interact is crucial to modern biology, with applications in drug discovery and protein design. Recent machine learning methods have formulated protein-small molecule docking as a generative problem with significant performance boosts over both traditional and deep learning baselines. In this work, we propose a similar approach for rigid protein-protein docking: DiffDock-PP is a diffusion generative model that learns to translate and rotate unbound protein structures into their bound conformations. We achieve state-of-the-art performance on DIPS with a median C-RMSD of 4.85, outperforming all considered baselines. Additionally, DiffDock-PP is faster than all search-based methods and generates reliable confidence estimates for its predictions. Our code is publicly available at $\texttt{https://github.com/ketatam/DiffDock-PP}$
RAFaRe: Learning Robust and Accurate Non-parametric 3D Face Reconstruction from Pseudo 2D&3D Pairs
Feb 10, 2023



Abstract:We propose a robust and accurate non-parametric method for single-view 3D face reconstruction (SVFR). While tremendous efforts have been devoted to parametric SVFR, a visible gap still lies between the result 3D shape and the ground truth. We believe there are two major obstacles: 1) the representation of the parametric model is limited to a certain face database; 2) 2D images and 3D shapes in the fitted datasets are distinctly misaligned. To resolve these issues, a large-scale pseudo 2D\&3D dataset is created by first rendering the detailed 3D faces, then swapping the face in the wild images with the rendered face. These pseudo 2D&3D pairs are created from publicly available datasets which eliminate the gaps between 2D and 3D data while covering diverse appearances, poses, scenes, and illumination. We further propose a non-parametric scheme to learn a well-generalized SVFR model from the created dataset, and the proposed hierarchical signed distance function turns out to be effective in predicting middle-scale and small-scale 3D facial geometry. Our model outperforms previous methods on FaceScape-wild/lab and MICC benchmarks and is well generalized to various appearances, poses, expressions, and in-the-wild environments. The code is released at http://github.com/zhuhao-nju/rafare .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge