Russell Littman
Group Contrastive Learning for Weakly Paired Multimodal Data
Feb 03, 2026Abstract:We present GROOVE, a semi-supervised multi-modal representation learning approach for high-content perturbation data where samples across modalities are weakly paired through shared perturbation labels but lack direct correspondence. Our primary contribution is GroupCLIP, a novel group-level contrastive loss that bridges the gap between CLIP for paired cross-modal data and SupCon for uni-modal supervised contrastive learning, addressing a fundamental gap in contrastive learning for weakly-paired settings. We integrate GroupCLIP with an on-the-fly backtranslating autoencoder framework to encourage cross-modally entangled representations while maintaining group-level coherence within a shared latent space. Critically, we introduce a comprehensive combinatorial evaluation framework that systematically assesses representation learners across multiple optimal transport aligners, addressing key limitations in existing evaluation strategies. This framework includes novel simulations that systematically vary shared versus modality-specific perturbation effects enabling principled assessment of method robustness. Our combinatorial benchmarking reveals that there is not yet an aligner that uniformly dominates across settings or modality pairs. Across simulations and two real single-cell genetic perturbation datasets, GROOVE performs on par with or outperforms existing approaches for downstream cross-modal matching and imputation tasks. Our ablation studies demonstrate that GroupCLIP is the key component driving performance gains. These results highlight the importance of leveraging group-level constraints for effective multi-modal representation learning in scenarios where only weak pairing is available.
Contextualizing biological perturbation experiments through language
Feb 28, 2025
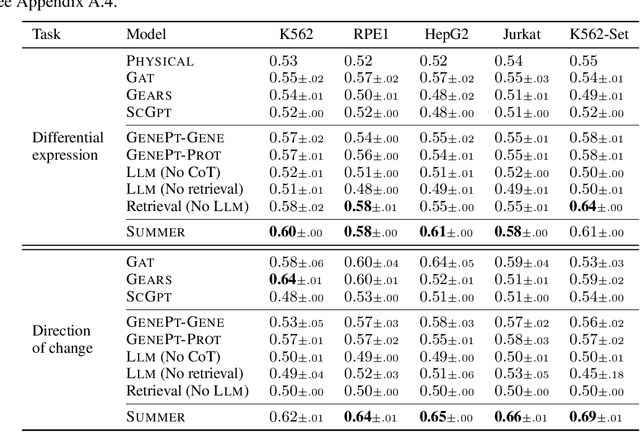

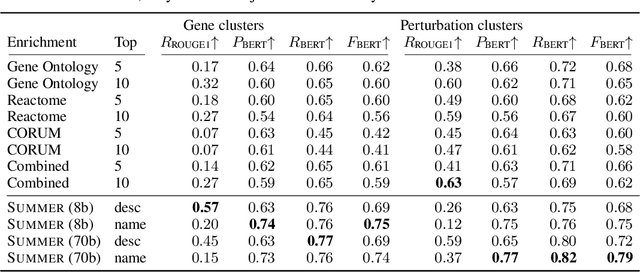
Abstract:High-content perturbation experiments allow scientists to probe biomolecular systems at unprecedented resolution, but experimental and analysis costs pose significant barriers to widespread adoption. Machine learning has the potential to guide efficient exploration of the perturbation space and extract novel insights from these data. However, current approaches neglect the semantic richness of the relevant biology, and their objectives are misaligned with downstream biological analyses. In this paper, we hypothesize that large language models (LLMs) present a natural medium for representing complex biological relationships and rationalizing experimental outcomes. We propose PerturbQA, a benchmark for structured reasoning over perturbation experiments. Unlike current benchmarks that primarily interrogate existing knowledge, PerturbQA is inspired by open problems in perturbation modeling: prediction of differential expression and change of direction for unseen perturbations, and gene set enrichment. We evaluate state-of-the-art machine learning and statistical approaches for modeling perturbations, as well as standard LLM reasoning strategies, and we find that current methods perform poorly on PerturbQA. As a proof of feasibility, we introduce Summer (SUMMarize, retrievE, and answeR, a simple, domain-informed LLM framework that matches or exceeds the current state-of-the-art. Our code and data are publicly available at https://github.com/genentech/PerturbQA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge