Tommi S. Jaakkola
Structural Constraint Integration in Generative Model for Discovery of Quantum Material Candidates
Jul 05, 2024Abstract:Billions of organic molecules are known, but only a tiny fraction of the functional inorganic materials have been discovered, a particularly relevant problem to the community searching for new quantum materials. Recent advancements in machine-learning-based generative models, particularly diffusion models, show great promise for generating new, stable materials. However, integrating geometric patterns into materials generation remains a challenge. Here, we introduce Structural Constraint Integration in the GENerative model (SCIGEN). Our approach can modify any trained generative diffusion model by strategic masking of the denoised structure with a diffused constrained structure prior to each diffusion step to steer the generation toward constrained outputs. Furthermore, we mathematically prove that SCIGEN effectively performs conditional sampling from the original distribution, which is crucial for generating stable constrained materials. We generate eight million compounds using Archimedean lattices as prototype constraints, with over 10% surviving a multi-staged stability pre-screening. High-throughput density functional theory (DFT) on 26,000 survived compounds shows that over 50% passed structural optimization at the DFT level. Since the properties of quantum materials are closely related to geometric patterns, our results indicate that SCIGEN provides a general framework for generating quantum materials candidates.
Improved motif-scaffolding with SE(3) flow matching
Jan 08, 2024Abstract:Protein design often begins with knowledge of a desired function from a motif which motif-scaffolding aims to construct a functional protein around. Recently, generative models have achieved breakthrough success in designing scaffolds for a diverse range of motifs. However, the generated scaffolds tend to lack structural diversity, which can hinder success in wet-lab validation. In this work, we extend FrameFlow, an SE(3) flow matching model for protein backbone generation, to perform motif-scaffolding with two complementary approaches. The first is motif amortization, in which FrameFlow is trained with the motif as input using a data augmentation strategy. The second is motif guidance, which performs scaffolding using an estimate of the conditional score from FrameFlow, and requires no additional training. Both approaches achieve an equivalent or higher success rate than previous state-of-the-art methods, with 2.5 times more structurally diverse scaffolds. Code: https://github.com/ microsoft/frame-flow.
Conformal Language Modeling
Jun 16, 2023Abstract:We propose a novel approach to conformal prediction for generative language models (LMs). Standard conformal prediction produces prediction sets -- in place of single predictions -- that have rigorous, statistical performance guarantees. LM responses are typically sampled from the model's predicted distribution over the large, combinatorial output space of natural language. Translating this process to conformal prediction, we calibrate a stopping rule for sampling different outputs from the LM that get added to a growing set of candidates until we are confident that the output set is sufficient. Since some samples may be low-quality, we also simultaneously calibrate and apply a rejection rule for removing candidates from the output set to reduce noise. Similar to conformal prediction, we prove that the sampled set returned by our procedure contains at least one acceptable answer with high probability, while still being empirically precise (i.e., small) on average. Furthermore, within this set of candidate responses, we show that we can also accurately identify subsets of individual components -- such as phrases or sentences -- that are each independently correct (e.g., that are not "hallucinations"), again with statistical guarantees. We demonstrate the promise of our approach on multiple tasks in open-domain question answering, text summarization, and radiology report generation using different LM variants.
DiffDock-PP: Rigid Protein-Protein Docking with Diffusion Models
Apr 08, 2023



Abstract:Understanding how proteins structurally interact is crucial to modern biology, with applications in drug discovery and protein design. Recent machine learning methods have formulated protein-small molecule docking as a generative problem with significant performance boosts over both traditional and deep learning baselines. In this work, we propose a similar approach for rigid protein-protein docking: DiffDock-PP is a diffusion generative model that learns to translate and rotate unbound protein structures into their bound conformations. We achieve state-of-the-art performance on DIPS with a median C-RMSD of 4.85, outperforming all considered baselines. Additionally, DiffDock-PP is faster than all search-based methods and generates reliable confidence estimates for its predictions. Our code is publicly available at $\texttt{https://github.com/ketatam/DiffDock-PP}$
PEOPL: Characterizing Privately Encoded Open Datasets with Public Labels
Mar 31, 2023Abstract:Allowing organizations to share their data for training of machine learning (ML) models without unintended information leakage is an open problem in practice. A promising technique for this still-open problem is to train models on the encoded data. Our approach, called Privately Encoded Open Datasets with Public Labels (PEOPL), uses a certain class of randomly constructed transforms to encode sensitive data. Organizations publish their randomly encoded data and associated raw labels for ML training, where training is done without knowledge of the encoding realization. We investigate several important aspects of this problem: We introduce information-theoretic scores for privacy and utility, which quantify the average performance of an unfaithful user (e.g., adversary) and a faithful user (e.g., model developer) that have access to the published encoded data. We then theoretically characterize primitives in building families of encoding schemes that motivate the use of random deep neural networks. Empirically, we compare the performance of our randomized encoding scheme and a linear scheme to a suite of computational attacks, and we also show that our scheme achieves competitive prediction accuracy to raw-sample baselines. Moreover, we demonstrate that multiple institutions, using independent random encoders, can collaborate to train improved ML models.
Adversarial Support Alignment
Mar 16, 2022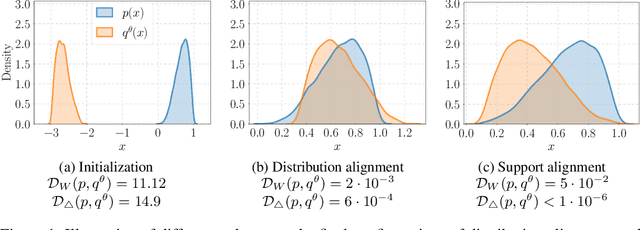
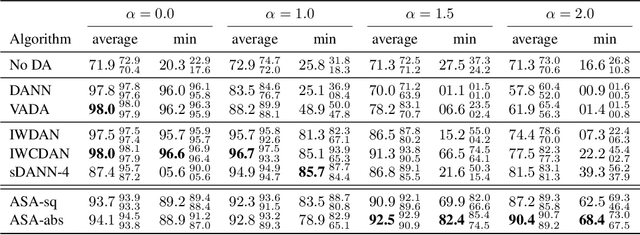
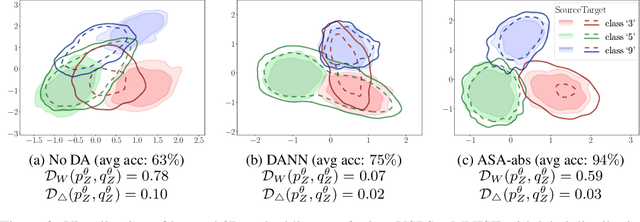
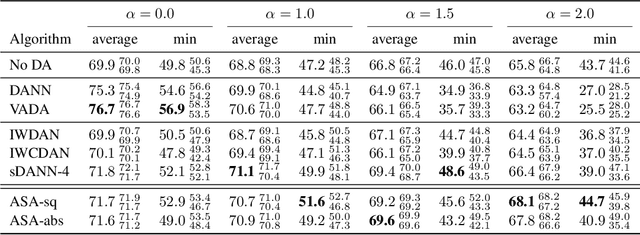
Abstract:We study the problem of aligning the supports of distributions. Compared to the existing work on distribution alignment, support alignment does not require the densities to be matched. We propose symmetric support difference as a divergence measure to quantify the mismatch between supports. We show that select discriminators (e.g. discriminator trained for Jensen-Shannon divergence) are able to map support differences as support differences in their one-dimensional output space. Following this result, our method aligns supports by minimizing a symmetrized relaxed optimal transport cost in the discriminator 1D space via an adversarial process. Furthermore, we show that our approach can be viewed as a limit of existing notions of alignment by increasing transportation assignment tolerance. We quantitatively evaluate the method across domain adaptation tasks with shifts in label distributions. Our experiments show that the proposed method is more robust against these shifts than other alignment-based baselines.
Syfer: Neural Obfuscation for Private Data Release
Jan 28, 2022Abstract:Balancing privacy and predictive utility remains a central challenge for machine learning in healthcare. In this paper, we develop Syfer, a neural obfuscation method to protect against re-identification attacks. Syfer composes trained layers with random neural networks to encode the original data (e.g. X-rays) while maintaining the ability to predict diagnoses from the encoded data. The randomness in the encoder acts as the private key for the data owner. We quantify privacy as the number of attacker guesses required to re-identify a single image (guesswork). We propose a contrastive learning algorithm to estimate guesswork. We show empirically that differentially private methods, such as DP-Image, obtain privacy at a significant loss of utility. In contrast, Syfer achieves strong privacy while preserving utility. For example, X-ray classifiers built with DP-image, Syfer, and original data achieve average AUCs of 0.53, 0.78, and 0.86, respectively.
Understanding Interlocking Dynamics of Cooperative Rationalization
Oct 26, 2021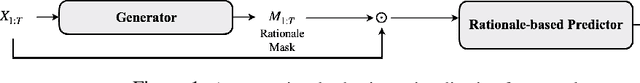
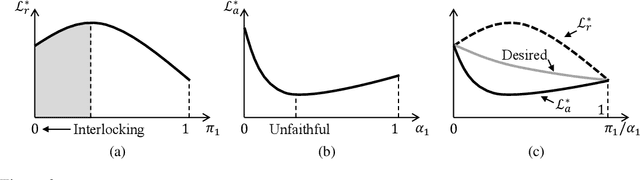
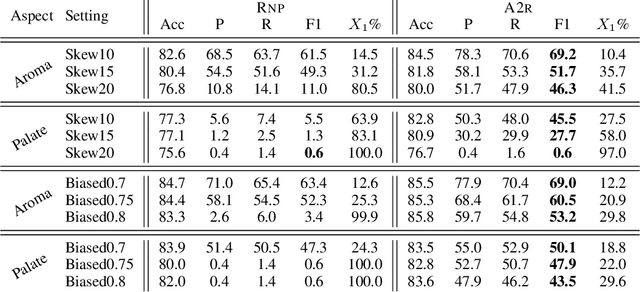
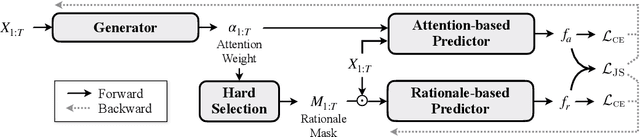
Abstract:Selective rationalization explains the prediction of complex neural networks by finding a small subset of the input that is sufficient to predict the neural model output. The selection mechanism is commonly integrated into the model itself by specifying a two-component cascaded system consisting of a rationale generator, which makes a binary selection of the input features (which is the rationale), and a predictor, which predicts the output based only on the selected features. The components are trained jointly to optimize prediction performance. In this paper, we reveal a major problem with such cooperative rationalization paradigm -- model interlocking. Interlocking arises when the predictor overfits to the features selected by the generator thus reinforcing the generator's selection even if the selected rationales are sub-optimal. The fundamental cause of the interlocking problem is that the rationalization objective to be minimized is concave with respect to the generator's selection policy. We propose a new rationalization framework, called A2R, which introduces a third component into the architecture, a predictor driven by soft attention as opposed to selection. The generator now realizes both soft and hard attention over the features and these are fed into the two different predictors. While the generator still seeks to support the original predictor performance, it also minimizes a gap between the two predictors. As we will show theoretically, since the attention-based predictor exhibits a better convexity property, A2R can overcome the concavity barrier. Our experiments on two synthetic benchmarks and two real datasets demonstrate that A2R can significantly alleviate the interlock problem and find explanations that better align with human judgments. We release our code at https://github.com/Gorov/Understanding_Interlocking.
GeoMol: Torsional Geometric Generation of Molecular 3D Conformer Ensembles
Jun 08, 2021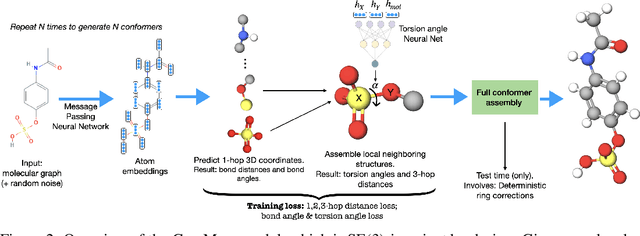
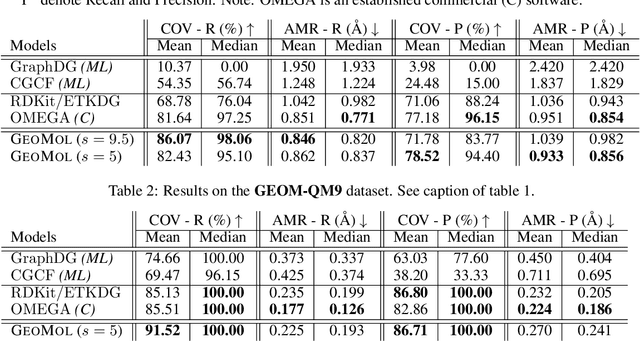
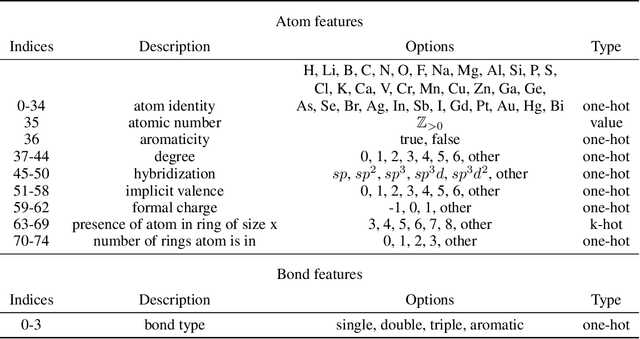
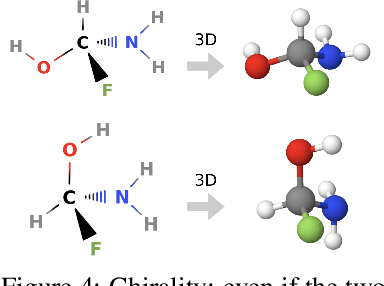
Abstract:Prediction of a molecule's 3D conformer ensemble from the molecular graph holds a key role in areas of cheminformatics and drug discovery. Existing generative models have several drawbacks including lack of modeling important molecular geometry elements (e.g. torsion angles), separate optimization stages prone to error accumulation, and the need for structure fine-tuning based on approximate classical force-fields or computationally expensive methods such as metadynamics with approximate quantum mechanics calculations at each geometry. We propose GeoMol--an end-to-end, non-autoregressive and SE(3)-invariant machine learning approach to generate distributions of low-energy molecular 3D conformers. Leveraging the power of message passing neural networks (MPNNs) to capture local and global graph information, we predict local atomic 3D structures and torsion angles, avoiding unnecessary over-parameterization of the geometric degrees of freedom (e.g. one angle per non-terminal bond). Such local predictions suffice both for the training loss computation, as well as for the full deterministic conformer assembly (at test time). We devise a non-adversarial optimal transport based loss function to promote diverse conformer generation. GeoMol predominantly outperforms popular open-source, commercial, or state-of-the-art machine learning (ML) models, while achieving significant speed-ups. We expect such differentiable 3D structure generators to significantly impact molecular modeling and related applications.
NeuraCrypt: Hiding Private Health Data via Random Neural Networks for Public Training
Jun 04, 2021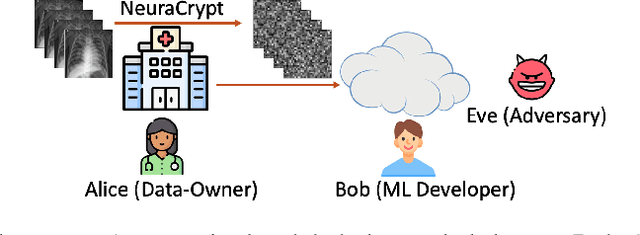
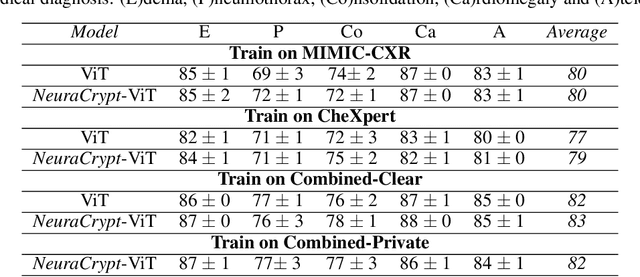
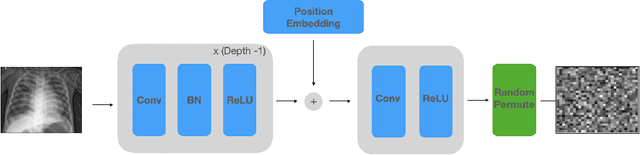

Abstract:Balancing the needs of data privacy and predictive utility is a central challenge for machine learning in healthcare. In particular, privacy concerns have led to a dearth of public datasets, complicated the construction of multi-hospital cohorts and limited the utilization of external machine learning resources. To remedy this, new methods are required to enable data owners, such as hospitals, to share their datasets publicly, while preserving both patient privacy and modeling utility. We propose NeuraCrypt, a private encoding scheme based on random deep neural networks. NeuraCrypt encodes raw patient data using a randomly constructed neural network known only to the data-owner, and publishes both the encoded data and associated labels publicly. From a theoretical perspective, we demonstrate that sampling from a sufficiently rich family of encoding functions offers a well-defined and meaningful notion of privacy against a computationally unbounded adversary with full knowledge of the underlying data-distribution. We propose to approximate this family of encoding functions through random deep neural networks. Empirically, we demonstrate the robustness of our encoding to a suite of adversarial attacks and show that NeuraCrypt achieves competitive accuracy to non-private baselines on a variety of x-ray tasks. Moreover, we demonstrate that multiple hospitals, using independent private encoders, can collaborate to train improved x-ray models. Finally, we release a challenge dataset to encourage the development of new attacks on NeuraCrypt.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge