Burkhard Hoeckendorf
Supervised Contrastive Block Disentanglement
Feb 11, 2025



Abstract:Real-world datasets often combine data collected under different experimental conditions. This yields larger datasets, but also introduces spurious correlations that make it difficult to model the phenomena of interest. We address this by learning two embeddings to independently represent the phenomena of interest and the spurious correlations. The embedding representing the phenomena of interest is correlated with the target variable $y$, and is invariant to the environment variable $e$. In contrast, the embedding representing the spurious correlations is correlated with $e$. The invariance to $e$ is difficult to achieve on real-world datasets. Our primary contribution is an algorithm called Supervised Contrastive Block Disentanglement (SCBD) that effectively enforces this invariance. It is based purely on Supervised Contrastive Learning, and applies to real-world data better than existing approaches. We empirically validate SCBD on two challenging problems. The first problem is domain generalization, where we achieve strong performance on a synthetic dataset, as well as on Camelyon17-WILDS. We introduce a single hyperparameter $\alpha$ to control the degree of invariance to $e$. When we increase $\alpha$ to strengthen the degree of invariance, out-of-distribution performance improves at the expense of in-distribution performance. The second problem is batch correction, in which we apply SCBD to preserve biological signal and remove inter-well batch effects when modeling single-cell perturbations from 26 million Optical Pooled Screening images.
Weakly Supervised Set-Consistency Learning Improves Morphological Profiling of Single-Cell Images
Jun 08, 2024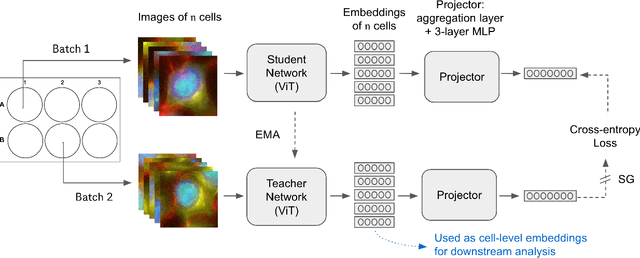



Abstract:Optical Pooled Screening (OPS) is a powerful tool combining high-content microscopy with genetic engineering to investigate gene function in disease. The characterization of high-content images remains an active area of research and is currently undergoing rapid innovation through the application of self-supervised learning and vision transformers. In this study, we propose a set-level consistency learning algorithm, Set-DINO, that combines self-supervised learning with weak supervision to improve learned representations of perturbation effects in single-cell images. Our method leverages the replicate structure of OPS experiments (i.e., cells undergoing the same genetic perturbation, both within and across batches) as a form of weak supervision. We conduct extensive experiments on a large-scale OPS dataset with more than 5000 genetic perturbations, and demonstrate that Set-DINO helps mitigate the impact of confounders and encodes more biologically meaningful information. In particular, Set-DINO recalls known biological relationships with higher accuracy compared to commonly used methods for morphological profiling, suggesting that it can generate more reliable insights from drug target discovery campaigns leveraging OPS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge