Kayvan Najarian
A Causal Machine Learning Framework for Treatment Personalization in Clinical Trials: Application to Ulcerative Colitis
Feb 09, 2026Abstract:Randomized controlled trials estimate average treatment effects, but treatment response heterogeneity motivates personalized approaches. A critical question is whether statistically detectable heterogeneity translates into improved treatment decisions -- these are distinct questions that can yield contradictory answers. We present a modular causal machine learning framework that evaluates each question separately: permutation importance identifies which features predict heterogeneity, best linear predictor (BLP) testing assesses statistical significance, and doubly robust policy evaluation measures whether acting on the heterogeneity improves patient outcomes. We apply this framework to patient-level data from the UNIFI maintenance trial of ustekinumab in ulcerative colitis, comparing placebo, standard-dose ustekinumab every 12 weeks, and dose-intensified ustekinumab every 8 weeks, using cross-fitted X-learner models with baseline demographics, medication history, week-8 clinical scores, laboratory biomarkers, and video-derived endoscopic features. BLP testing identified strong associations between endoscopic features and treatment effect heterogeneity for ustekinumab versus placebo, yet doubly robust policy evaluation showed no improvement in expected remission from incorporating endoscopic features, and out-of-fold multi-arm evaluation showed worse performance. Diagnostic comparison of prognostic contribution against policy value revealed that endoscopic scores behaved as disease severity markers -- improving outcome prediction in untreated patients but adding noise to treatment selection -- while clinical variables (fecal calprotectin, age, CRP) captured the decision-relevant variation. These results demonstrate that causal machine learning applications to clinical trials should include policy-level evaluation alongside heterogeneity testing.
Rethinking the long-range dependency in Mamba/SSM and transformer models
Sep 04, 2025Abstract:Long-range dependency is one of the most desired properties of recent sequence models such as state-space models (particularly Mamba) and transformer models. New model architectures are being actively developed and benchmarked for prediction tasks requiring long-range dependency. However, the capability of modeling long-range dependencies of these models has not been investigated from a theoretical perspective, which hinders a systematic improvement on this aspect. In this work, we mathematically define long-range dependency using the derivative of hidden states with respect to past inputs and compare the capability of SSM and transformer models of modeling long-range dependency based on this definition. We showed that the long-range dependency of SSM decays exponentially with the sequence length, which aligns with the exponential decay of memory function in RNN. But the attention mechanism used in transformers is more flexible and is not constrained to exponential decay, which could in theory perform better at modeling long-range dependency with sufficient training data, computing resources, and proper training. To combine the flexibility of long-range dependency of attention mechanism and computation efficiency of SSM, we propose a new formulation for hidden state update in SSM and prove its stability under a standard Gaussian distribution of the input data.
Tensor Denoising via Amplification and Stable Rank Methods
Jan 10, 2023



Abstract:Tensors in the form of multilinear arrays are ubiquitous in data science applications. Captured real-world data, including video, hyperspectral images, and discretized physical systems, naturally occur as tensors and often come with attendant noise. Under the additive noise model and with the assumption that the underlying clean tensor has low rank, many denoising methods have been created that utilize tensor decomposition to effect denoising through low rank tensor approximation. However, all such decomposition methods require estimating the tensor rank, or related measures such as the tensor spectral and nuclear norms, all of which are NP-hard problems. In this work we adapt the previously developed framework of tensor amplification, which provides good approximations of the spectral and nuclear tensor norms, to denoising synthetic tensors of various sizes, ranks, and noise levels, along with real-world tensors derived from physiological signals. We also introduce denoising methods based on two variations of rank estimates called stable $X$-rank and stable slice rank. The experimental results show that in the low rank context, tensor-based amplification provides comparable denoising performance in high signal-to-noise ratio (SNR) settings and superior performance in noisy (i.e., low SNR) settings, while the stable $X$-rank method achieves superior denoising performance on the physiological signal data.
Prediction of Oral Food Challenges via Machine Learning
Aug 17, 2022
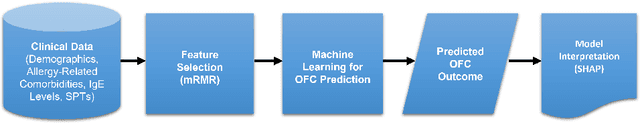
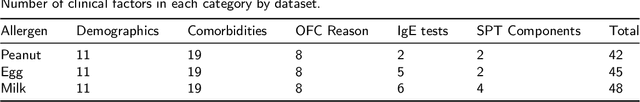
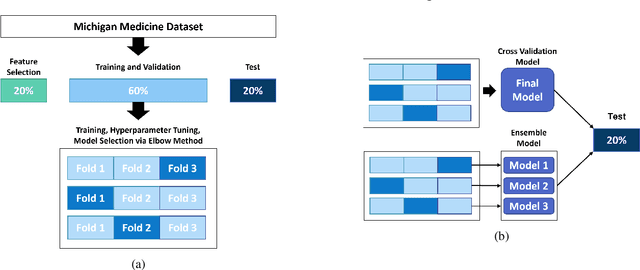
Abstract:Oral Food Challenges (OFCs) are essential to accurately diagnosing food allergy in patients. However, patients are hesitant to undergo OFCs, and for those that do, there is limited access to allergists in rural/community healthcare settings. The prediction of OFC outcomes through machine learning methods can facilitate the de-labeling of food allergens at home, improve patient and physician comfort during OFCs, and economize medical resources by minimizing the number of OFCs performed. Clinical data was gathered from 1,112 patients who collectively underwent a total of 1,284 OFCs, and consisted of clinical factors including serum specific IgE, total IgE, skin prick tests (SPTs), symptoms, sex, and age. Using these clinical features, machine learning models were constructed to predict outcomes for peanut, egg, and milk challenge. The best performing model for each allergen was created using the Learning Using Concave and Convex Kernels (LUCCK) method, which achieved an Area under the Curve (AUC) for peanut, egg, and milk OFC prediction of 0.76, 0.68, and 0.70, respectively. Model interpretation via SHapley Additive exPlanations (SHAP) indicate that specific IgE, along with wheal and flare values from SPTs, are highly predictive of OFC outcomes. The results of this analysis suggest that machine learning has the potential to predict OFC outcomes and reveal relevant clinical factors for further study.
A Novel Tropical Geometry-based Interpretable Machine Learning Method: Application in Prognosis of Advanced Heart Failure
Dec 09, 2021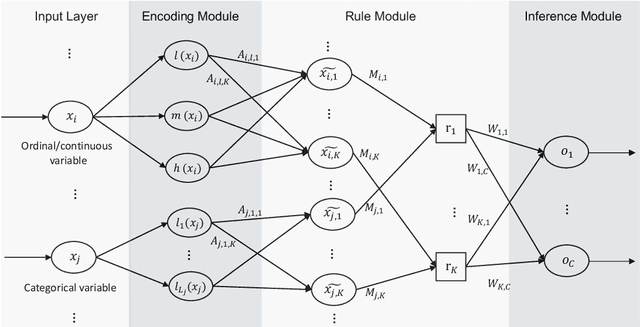
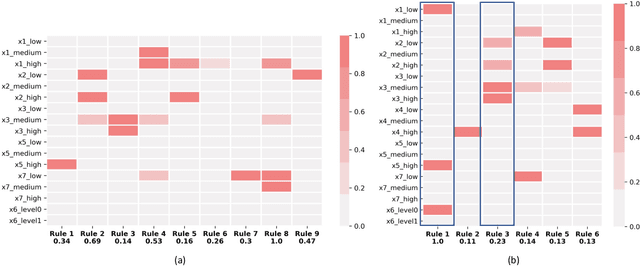
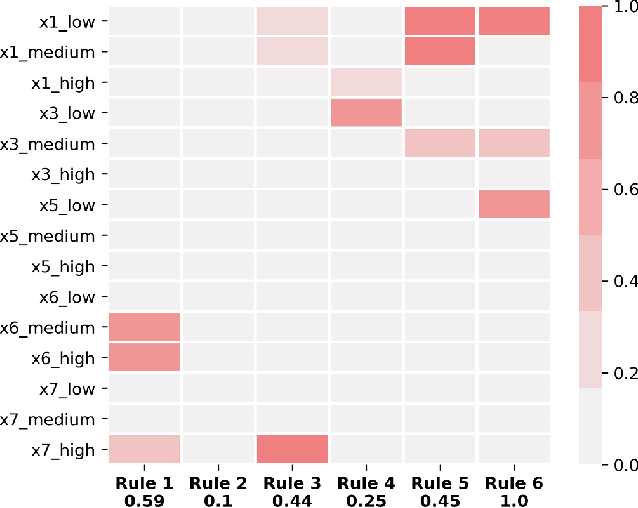
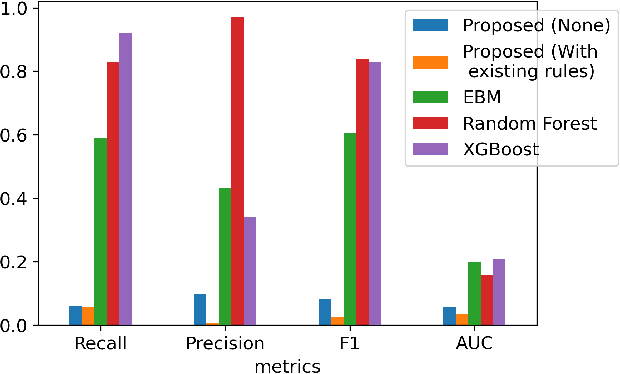
Abstract:A model's interpretability is essential to many practical applications such as clinical decision support systems. In this paper, a novel interpretable machine learning method is presented, which can model the relationship between input variables and responses in humanly understandable rules. The method is built by applying tropical geometry to fuzzy inference systems, wherein variable encoding functions and salient rules can be discovered by supervised learning. Experiments using synthetic datasets were conducted to investigate the performance and capacity of the proposed algorithm in classification and rule discovery. Furthermore, the proposed method was applied to a clinical application that identified heart failure patients that would benefit from advanced therapies such as heart transplant or durable mechanical circulatory support. Experimental results show that the proposed network achieved great performance on the classification tasks. In addition to learning humanly understandable rules from the dataset, existing fuzzy domain knowledge can be easily transferred into the network and used to facilitate model training. From our results, the proposed model and the ability of learning existing domain knowledge can significantly improve the model generalizability. The characteristics of the proposed network make it promising in applications requiring model reliability and justification.
Motion-based Camera Localization System in Colonoscopy Videos
Dec 04, 2020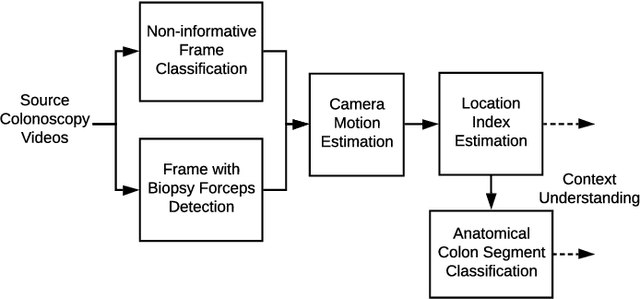
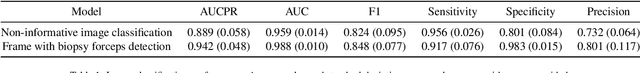
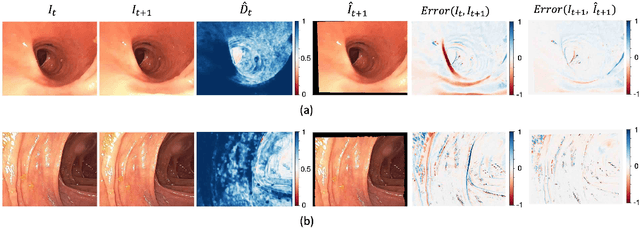
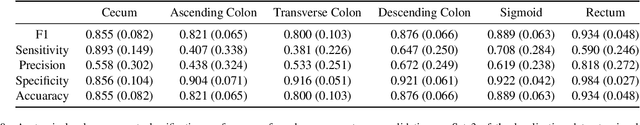
Abstract:Optical colonoscopy is an essential diagnostic and prognostic tool for many gastrointestinal diseases including cancer screening and staging, intestinal bleeding, diarrhea, abdominal symptom evaluation, and inflammatory bowel disease assessment. However, the evaluation, classification, and quantification of findings on colonoscopy are subject to inter-observer variation. Automated assessment of colonoscopy is of interest considering the subjectivity present in qualitative human interpretations of colonoscopy findings. Localization of the camera is an essential element to consider when inferring the meaning and context of findings for diseases evaluated by colonoscopy. In this study, we proposed a camera localization system to estimate the approximate anatomic location of the camera and classify the anatomical colon segment the camera is in. The camera localization system starts with non-informative frame detection to remove frames without camera motion information. Then a self-training end-to-end convolutional neural network was built to estimate the camera motion. With the estimated camera motion, the camera trajectory can be derived, and the location index can be calculated. Based on the estimated location index, anatomical colon segment classification was performed by building the colon template. The algorithm was trained and validated using colonoscopy videos collected from routine clinical practice. From our results, the average accuracy of the classification is 0.759, which is substantially higher than the performance of using the location index built from other methods.
Radial basis function kernel optimization for Support Vector Machine classifiers
Jul 16, 2020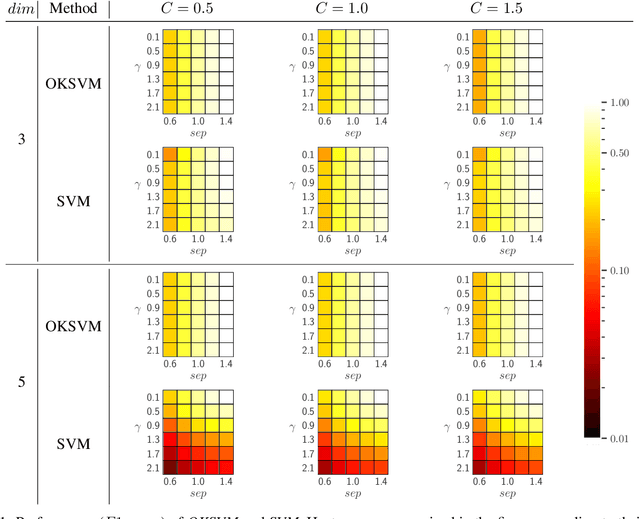
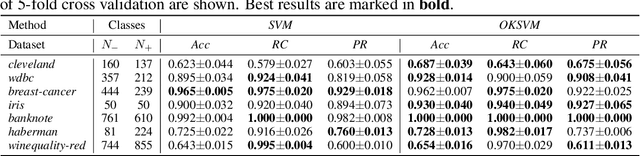

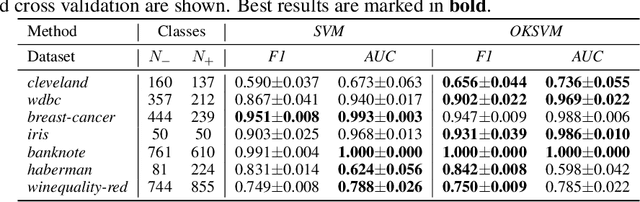
Abstract:Support Vector Machines (SVMs) are still one of the most popular and precise classifiers. The Radial Basis Function (RBF) kernel has been used in SVMs to separate among classes with considerable success. However, there is an intrinsic dependence on the initial value of the kernel hyperparameter. In this work, we propose OKSVM, an algorithm that automatically learns the RBF kernel hyperparameter and adjusts the SVM weights simultaneously. The proposed optimization technique is based on a gradient descent method. We analyze the performance of our approach with respect to the classical SVM for classification on synthetic and real data. Experimental results show that OKSVM performs better irrespective of the initial values of the RBF hyperparameter.
Fetal Ultrasound Image Segmentation for Measuring Biometric Parameters Using Multi-Task Deep Learning
Aug 31, 2019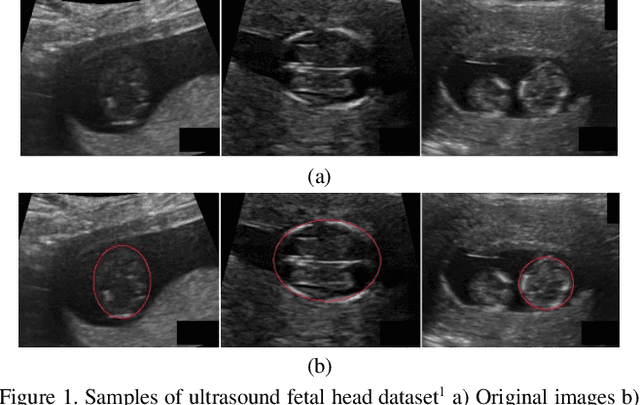
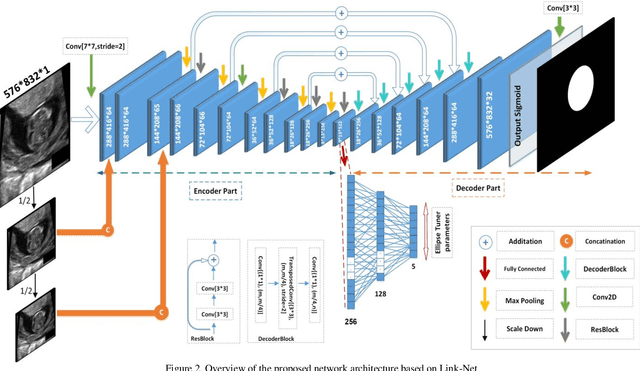

Abstract:Ultrasound imaging is a standard examination during pregnancy that can be used for measuring specific biometric parameters towards prenatal diagnosis and estimating gestational age. Fetal head circumference (HC) is one of the significant factors to determine the fetus growth and health. In this paper, a multi-task deep convolutional neural network is proposed for automatic segmentation and estimation of HC ellipse by minimizing a compound cost function composed of segmentation dice score and MSE of ellipse parameters. Experimental results on fetus ultrasound dataset in different trimesters of pregnancy show that the segmentation results and the extracted HC match well with the radiologist annotations. The obtained dice scores of the fetal head segmentation and the accuracy of HC evaluations are comparable to the state-of-the-art.
Gland Segmentation in Histopathology Images Using Deep Networks and Handcrafted Features
Aug 31, 2019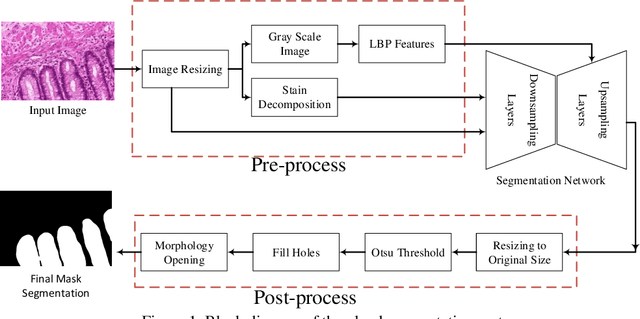
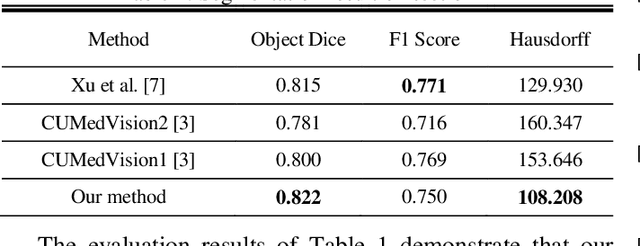

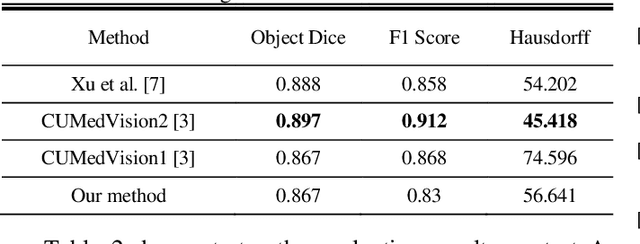
Abstract:Histopathology images contain essential information for medical diagnosis and prognosis of cancerous disease. Segmentation of glands in histopathology images is a primary step for analysis and diagnosis of an unhealthy patient. Due to the widespread application and the great success of deep neural networks in intelligent medical diagnosis and histopathology, we propose a modified version of LinkNet for gland segmentation and recognition of malignant cases. We show that using specific handcrafted features such as invariant local binary pattern drastically improves the system performance. The experimental results demonstrate the competency of the proposed system against state-of-the-art methods. We achieved the best results in testing on section B images of the Warwick-QU dataset and obtained comparable results on section A images.
An Unsupervised Feature Learning Approach to Reduce False Alarm Rate in ICUs
Apr 17, 2019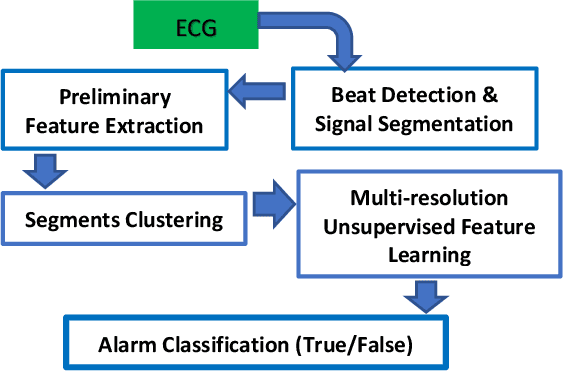
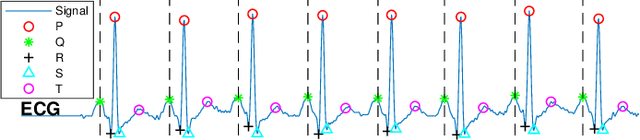
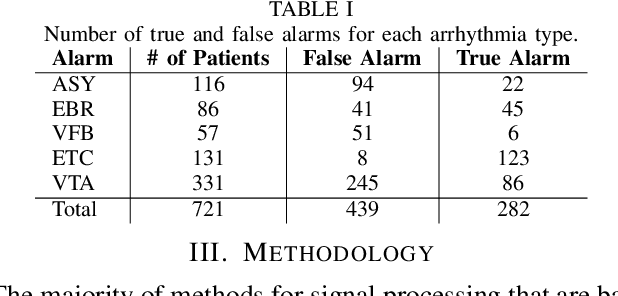
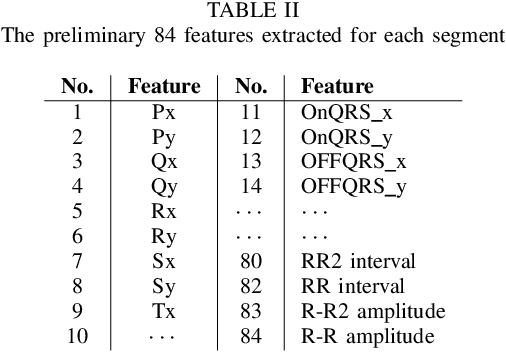
Abstract:The high rate of false alarms in intensive care units (ICUs) is one of the top challenges of using medical technology in hospitals. These false alarms are often caused by patients' movements, detachment of monitoring sensors, or different sources of noise and interference that impact the collected signals from different monitoring devices. In this paper, we propose a novel set of high-level features based on unsupervised feature learning technique in order to effectively capture the characteristics of different arrhythmia in electrocardiogram (ECG) signal and differentiate them from irregularity in signals due to different sources of signal disturbances. This unsupervised feature learning technique, first extracts a set of low-level features from all existing heart cycles of a patient, and then clusters these segments for each individual patient to provide a set of prominent high-level features. The objective of the clustering phase is to enable the classification method to differentiate between the high-level features extracted from normal and abnormal cycles (i.e., either due to arrhythmia or different sources of distortions in signal) in order to put more attention to the features extracted from abnormal portion of the signal that contribute to the alarm. The performance of this method is evaluated using the 2015 PhysioNet/Computing in Cardiology Challenge dataset for reducing false arrhythmia alarms in the ICUs. As confirmed by the experimental results, the proposed method offers a considerable performance in terms of accuracy, sensitivity and specificity of alarm detection only using a few high-level features that are extracted from one single lead ECG signal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge