Aakash Kaku
Quantifying Impairment and Disease Severity Using AI Models Trained on Healthy Subjects
Nov 21, 2023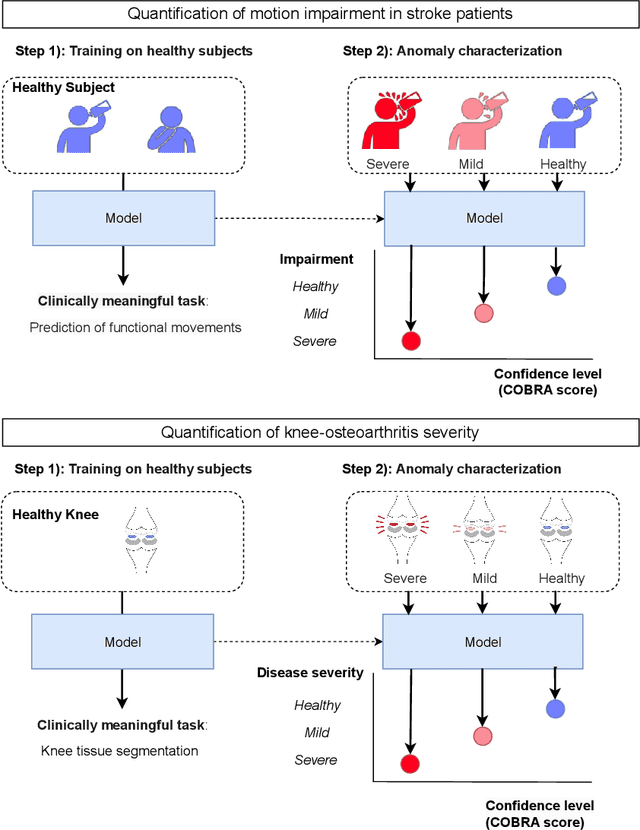
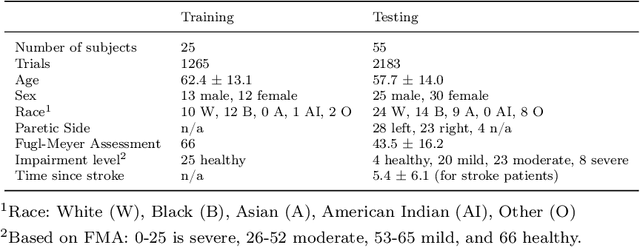
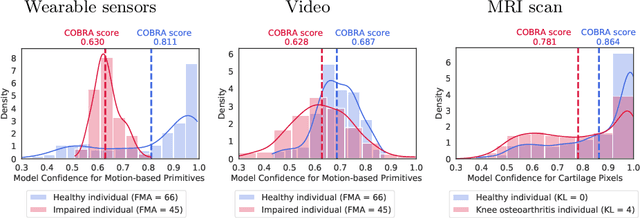
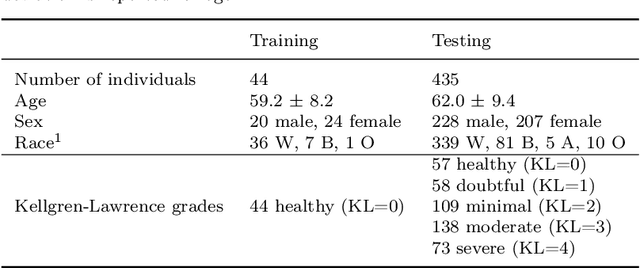
Abstract:Automatic assessment of impairment and disease severity is a key challenge in data-driven medicine. We propose a novel framework to address this challenge, which leverages AI models trained exclusively on healthy individuals. The COnfidence-Based chaRacterization of Anomalies (COBRA) score exploits the decrease in confidence of these models when presented with impaired or diseased patients to quantify their deviation from the healthy population. We applied the COBRA score to address a key limitation of current clinical evaluation of upper-body impairment in stroke patients. The gold-standard Fugl-Meyer Assessment (FMA) requires in-person administration by a trained assessor for 30-45 minutes, which restricts monitoring frequency and precludes physicians from adapting rehabilitation protocols to the progress of each patient. The COBRA score, computed automatically in under one minute, is shown to be strongly correlated with the FMA on an independent test cohort for two different data modalities: wearable sensors ($\rho = 0.845$, 95% CI [0.743,0.908]) and video ($\rho = 0.746$, 95% C.I [0.594, 0.847]). To demonstrate the generalizability of the approach to other conditions, the COBRA score was also applied to quantify severity of knee osteoarthritis from magnetic-resonance imaging scans, again achieving significant correlation with an independent clinical assessment ($\rho = 0.644$, 95% C.I [0.585,0.696]).
PrimSeq: a deep learning-based pipeline to quantitate rehabilitation training
Dec 22, 2021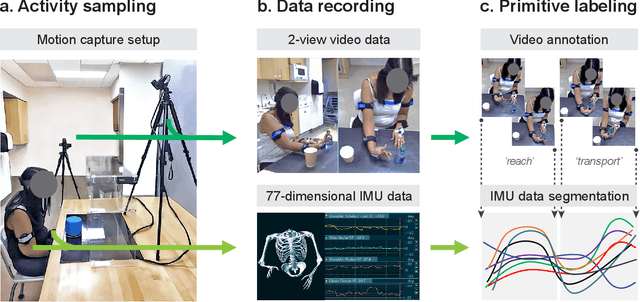
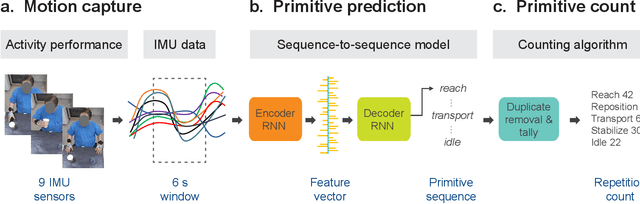
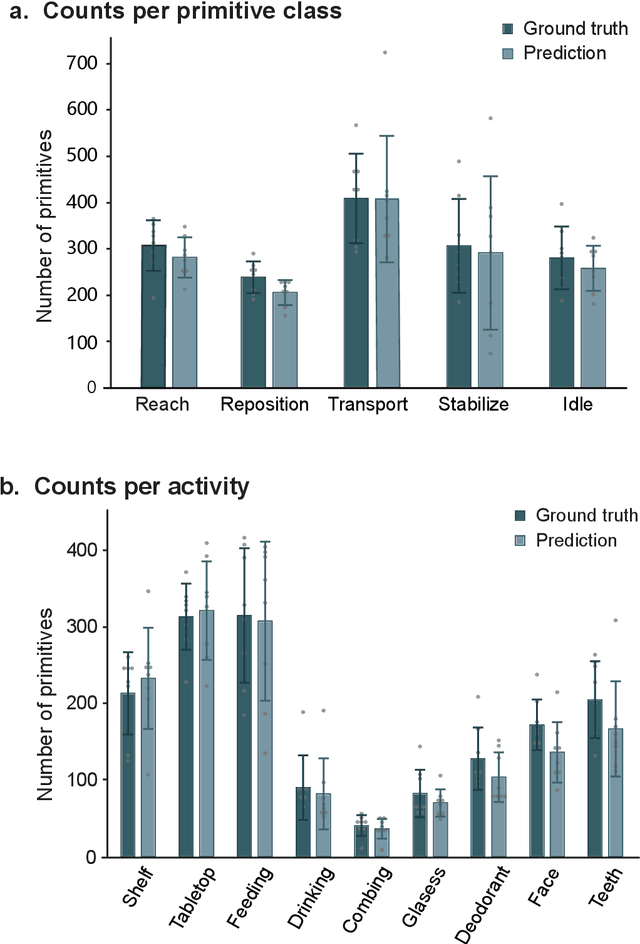
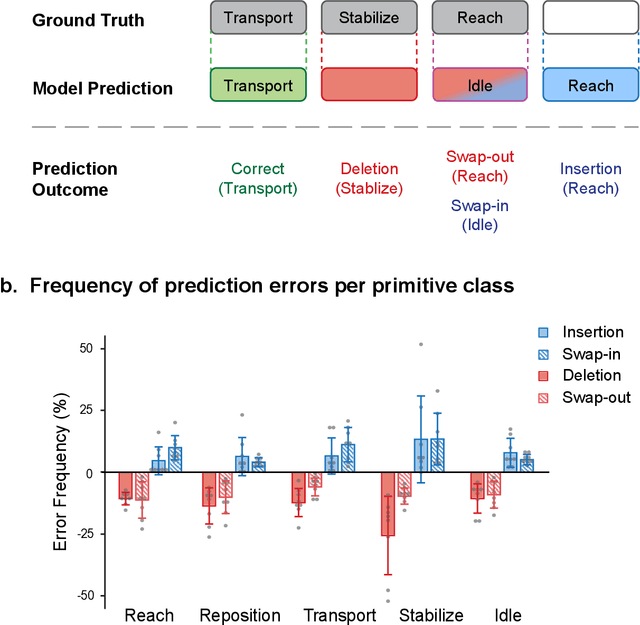
Abstract:Stroke rehabilitation seeks to increase neuroplasticity through the repeated practice of functional motions, but may have minimal impact on recovery because of insufficient repetitions. The optimal training content and quantity are currently unknown because no practical tools exist to measure them. Here, we present PrimSeq, a pipeline to classify and count functional motions trained in stroke rehabilitation. Our approach integrates wearable sensors to capture upper-body motion, a deep learning model to predict motion sequences, and an algorithm to tally motions. The trained model accurately decomposes rehabilitation activities into component functional motions, outperforming competitive machine learning methods. PrimSeq furthermore quantifies these motions at a fraction of the time and labor costs of human experts. We demonstrate the capabilities of PrimSeq in previously unseen stroke patients with a range of upper extremity motor impairment. We expect that these advances will support the rigorous measurement required for quantitative dosing trials in stroke rehabilitation.
Deep Probability Estimation
Nov 21, 2021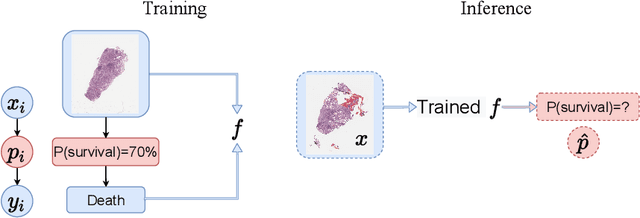
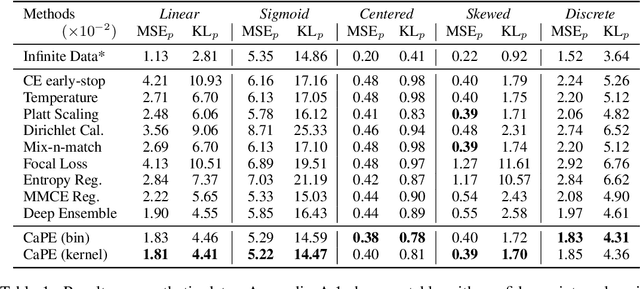
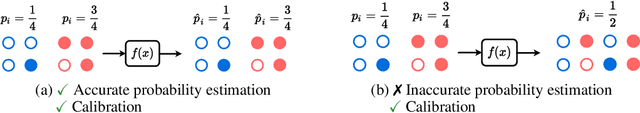
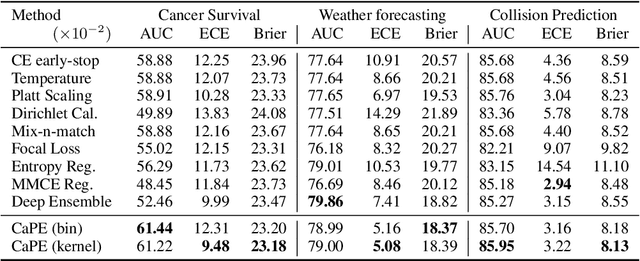
Abstract:Reliable probability estimation is of crucial importance in many real-world applications where there is inherent uncertainty, such as weather forecasting, medical prognosis, or collision avoidance in autonomous vehicles. Probability-estimation models are trained on observed outcomes (e.g. whether it has rained or not, or whether a patient has died or not), because the ground-truth probabilities of the events of interest are typically unknown. The problem is therefore analogous to binary classification, with the important difference that the objective is to estimate probabilities rather than predicting the specific outcome. The goal of this work is to investigate probability estimation from high-dimensional data using deep neural networks. There exist several methods to improve the probabilities generated by these models but they mostly focus on classification problems where the probabilities are related to model uncertainty. In the case of problems with inherent uncertainty, it is challenging to evaluate performance without access to ground-truth probabilities. To address this, we build a synthetic dataset to study and compare different computable metrics. We evaluate existing methods on the synthetic data as well as on three real-world probability estimation tasks, all of which involve inherent uncertainty: precipitation forecasting from radar images, predicting cancer patient survival from histopathology images, and predicting car crashes from dashcam videos. Finally, we also propose a new method for probability estimation using neural networks, which modifies the training process to promote output probabilities that are consistent with empirical probabilities computed from the data. The method outperforms existing approaches on most metrics on the simulated as well as real-world data.
Sequence-to-Sequence Modeling for Action Identification at High Temporal Resolution
Nov 03, 2021
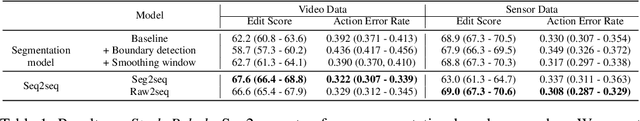
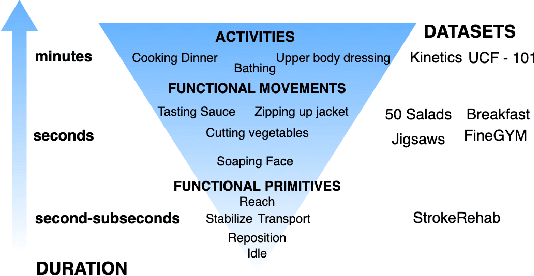
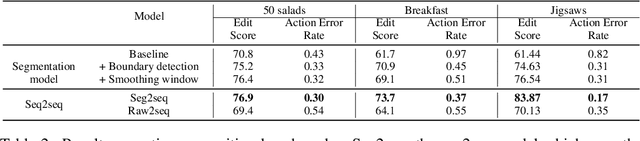
Abstract:Automatic action identification from video and kinematic data is an important machine learning problem with applications ranging from robotics to smart health. Most existing works focus on identifying coarse actions such as running, climbing, or cutting a vegetable, which have relatively long durations. This is an important limitation for applications that require the identification of subtle motions at high temporal resolution. For example, in stroke recovery, quantifying rehabilitation dose requires differentiating motions with sub-second durations. Our goal is to bridge this gap. To this end, we introduce a large-scale, multimodal dataset, StrokeRehab, as a new action-recognition benchmark that includes subtle short-duration actions labeled at a high temporal resolution. These short-duration actions are called functional primitives, and consist of reaches, transports, repositions, stabilizations, and idles. The dataset consists of high-quality Inertial Measurement Unit sensors and video data of 41 stroke-impaired patients performing activities of daily living like feeding, brushing teeth, etc. We show that current state-of-the-art models based on segmentation produce noisy predictions when applied to these data, which often leads to overcounting of actions. To address this, we propose a novel approach for high-resolution action identification, inspired by speech-recognition techniques, which is based on a sequence-to-sequence model that directly predicts the sequence of actions. This approach outperforms current state-of-the-art methods on the StrokeRehab dataset, as well as on the standard benchmark datasets 50Salads, Breakfast, and Jigsaws.
Intermediate Layers Matter in Momentum Contrastive Self Supervised Learning
Oct 27, 2021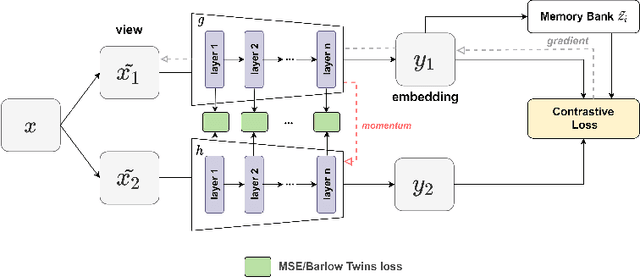
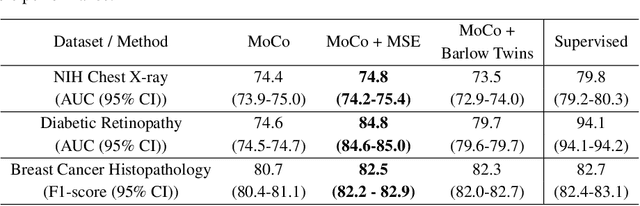
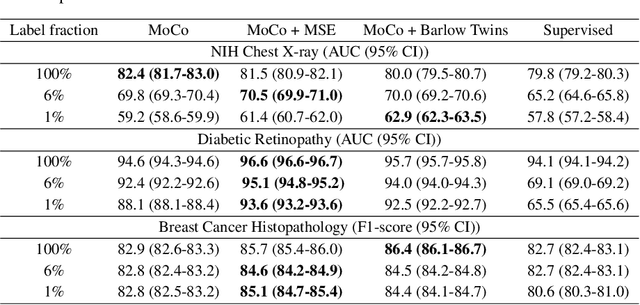
Abstract:We show that bringing intermediate layers' representations of two augmented versions of an image closer together in self-supervised learning helps to improve the momentum contrastive (MoCo) method. To this end, in addition to the contrastive loss, we minimize the mean squared error between the intermediate layer representations or make their cross-correlation matrix closer to an identity matrix. Both loss objectives either outperform standard MoCo, or achieve similar performances on three diverse medical imaging datasets: NIH-Chest Xrays, Breast Cancer Histopathology, and Diabetic Retinopathy. The gains of the improved MoCo are especially large in a low-labeled data regime (e.g. 1% labeled data) with an average gain of 5% across three datasets. We analyze the models trained using our novel approach via feature similarity analysis and layer-wise probing. Our analysis reveals that models trained via our approach have higher feature reuse compared to a standard MoCo and learn informative features earlier in the network. Finally, by comparing the output probability distribution of models fine-tuned on small versus large labeled data, we conclude that our proposed method of pre-training leads to lower Kolmogorov-Smirnov distance, as compared to a standard MoCo. This provides additional evidence that our proposed method learns more informative features in the pre-training phase which could be leveraged in a low-labeled data regime.
An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department
Aug 04, 2020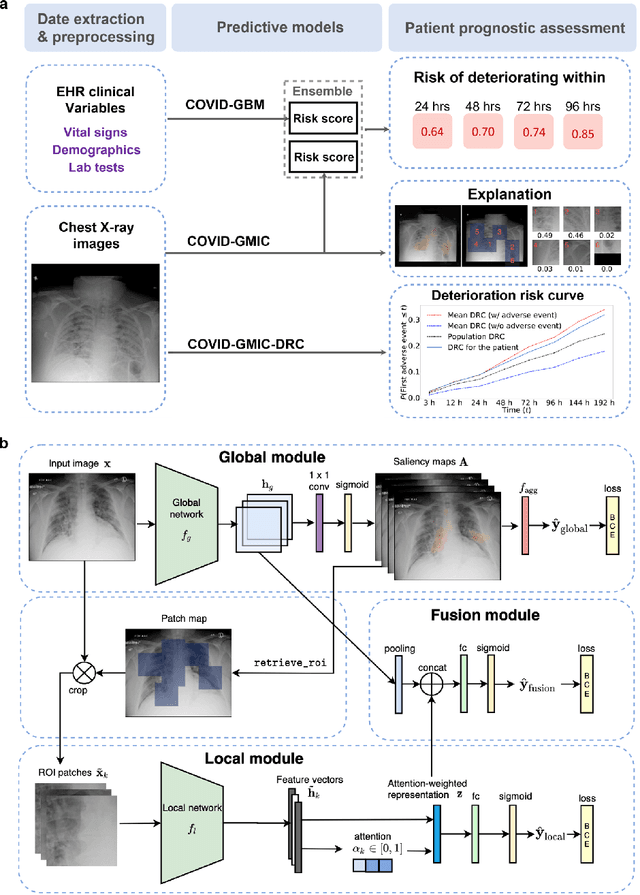
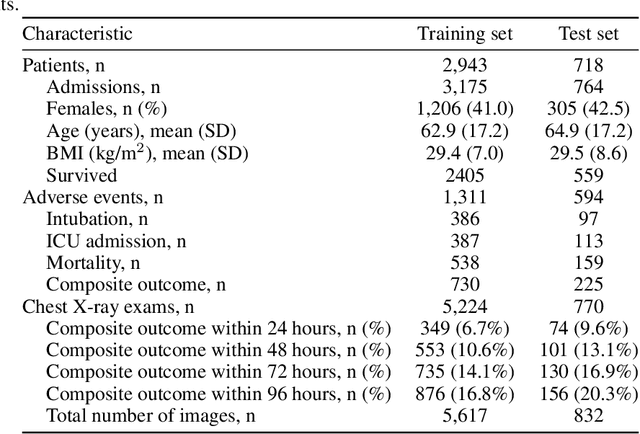
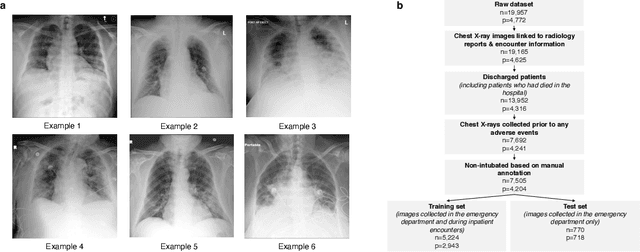
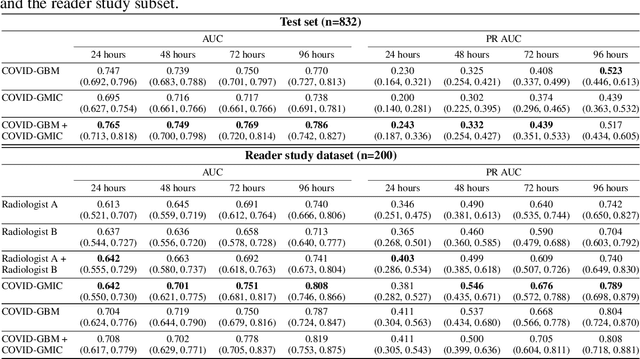
Abstract:During the COVID-19 pandemic, rapid and accurate triage of patients at the emergency department is critical to inform decision-making. We propose a data-driven approach for automatic prediction of deterioration risk using a deep neural network that learns from chest X-ray images, and a gradient boosting model that learns from routine clinical variables. Our AI prognosis system, trained using data from 3,661 patients, achieves an AUC of 0.786 (95% CI: 0.742-0.827) when predicting deterioration within 96 hours. The deep neural network extracts informative areas of chest X-ray images to assist clinicians in interpreting the predictions, and performs comparably to two radiologists in a reader study. In order to verify performance in a real clinical setting, we silently deployed a preliminary version of the deep neural network at NYU Langone Health during the first wave of the pandemic, which produced accurate predictions in real-time. In summary, our findings demonstrate the potential of the proposed system for assisting front-line physicians in the triage of COVID-19 patients.
Towards data-driven stroke rehabilitation via wearable sensors and deep learning
Apr 22, 2020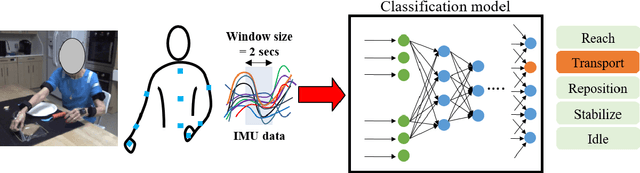
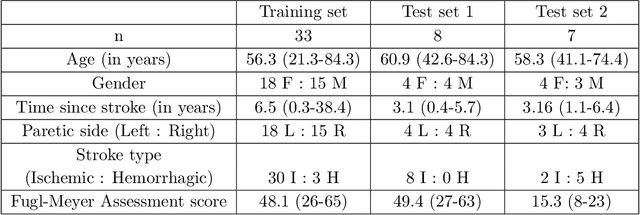
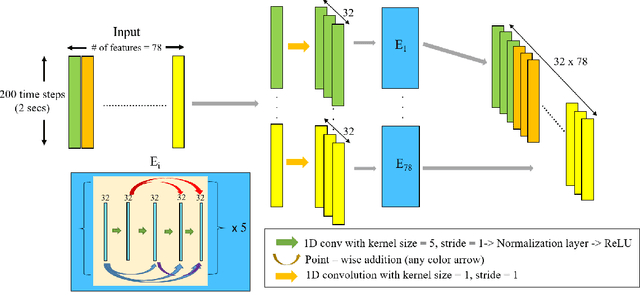
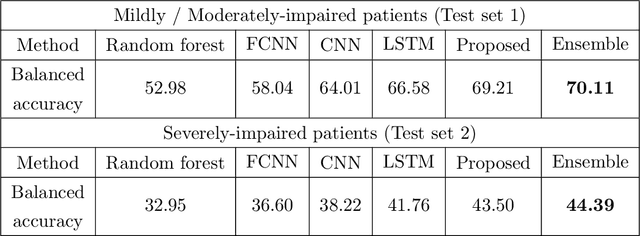
Abstract:Recovery after stroke is often incomplete, but rehabilitation training may potentiate recovery by engaging endogenous neuroplasticity. In preclinical models of stroke, high doses of rehabilitation training are required to restore functional movement to the affected limbs of animals. In humans, however, the necessary dose of training to potentiate recovery is not known. This ignorance stems from the lack of objective, pragmatic approaches for measuring training doses in rehabilitation activities. Here, to develop a measurement approach, we took the critical first step of automatically identifying functional primitives, the basic building block of activities. Forty-eight individuals with chronic stroke performed a variety of rehabilitation activities while wearing inertial measurement units (IMUs) to capture upper body motion. Primitives were identified by human labelers, who labeled and segmented the associated IMU data. We performed automatic classification of these primitives using machine learning. We designed a convolutional neural network model that outperformed existing methods. The model includes an initial module to compute separate embeddings of different physical quantities in the sensor data. In addition, it replaces batch normalization (which performs normalization based on statistics computed from the training data) with instance normalization (which uses statistics computed from the test data). This increases robustness to possible distributional shifts when applying the method to new patients. With this approach, we attained an average classification accuracy of 70%. Thus, using a combination of IMU-based motion capture and deep learning, we were able to identify primitives automatically. This approach builds towards objectively-measured rehabilitation training, enabling the identification and counting of functional primitives that accrues to a training dose.
Be Like Water: Robustness to Extraneous Variables Via Adaptive Feature Normalization
Feb 25, 2020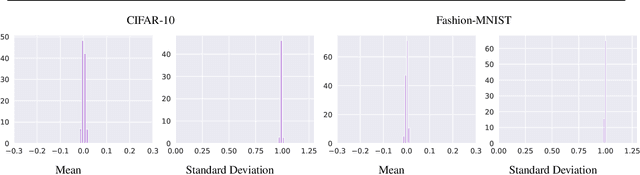
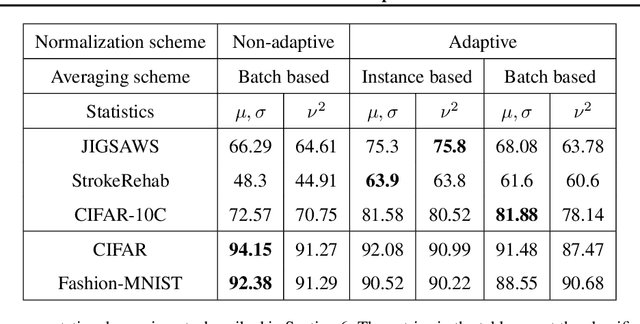
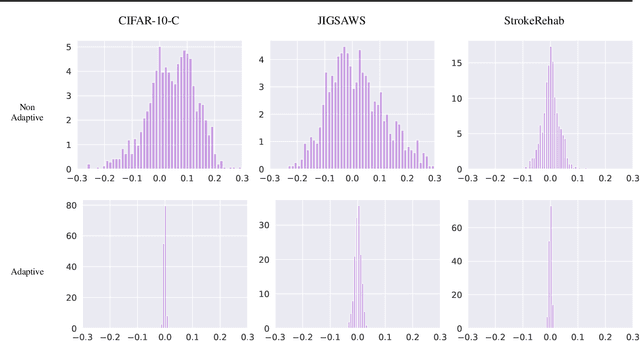
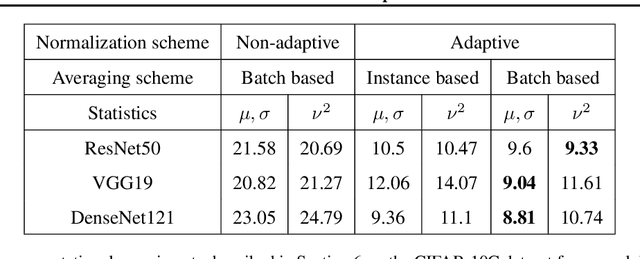
Abstract:Extraneous variables are variables that are irrelevant for a certain task, but heavily affect the distribution of the available data. In this work, we show that the presence of such variables can degrade the performance of deep-learning models. We study three datasets where there is a strong influence of known extraneous variables: classification of upper-body movements in stroke patients, annotation of surgical activities, and recognition of corrupted images. Models trained with batch normalization learn features that are highly dependent on the extraneous variables. In batch normalization, the statistics used to normalize the features are learned from the training set and fixed at test time, which produces a mismatch in the presence of varying extraneous variables. We demonstrate that estimating the feature statistics adaptively during inference, as in instance normalization, addresses this issue, producing normalized features that are more robust to changes in the extraneous variables. This results in a significant gain in performance for different network architectures and choices of feature statistics.
Knee Cartilage Segmentation Using Diffusion-Weighted MRI
Dec 04, 2019
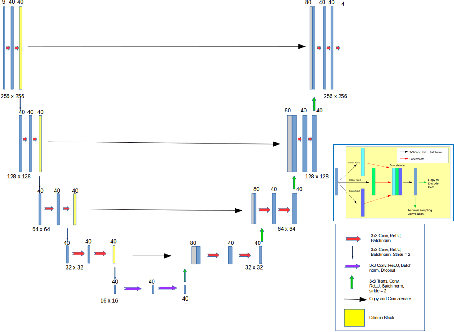
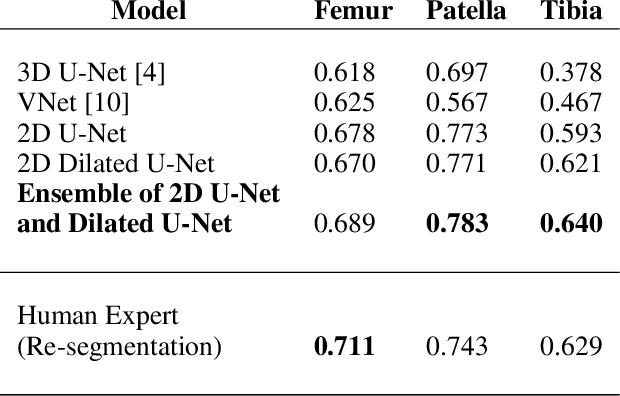
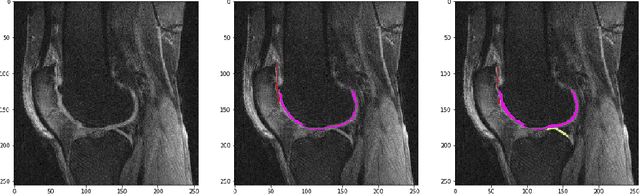
Abstract:The integrity of articular cartilage is a crucial aspect in the early diagnosis of osteoarthritis (OA). Many novel MRI techniques have the potential to assess compositional changes of the cartilage extracellular matrix. Among these techniques, diffusion tensor imaging (DTI) of cartilage provides a simultaneous assessment of the two principal components of the solid matrix: collagen structure and proteoglycan concentration. DTI, as for any other compositional MRI technique, require a human expert to perform segmentation manually. The manual segmentation is error-prone and time-consuming ($\sim$ few hours per subject). We use an ensemble of modified U-Nets to automate this segmentation task. We benchmark our model against a human expert test-retest segmentation and conclude that our model is superior for Patellar and Tibial cartilage using dice score as the comparison metric. In the end, we do a perturbation analysis to understand the sensitivity of our model to the different components of our input. We also provide confidence maps for the predictions so that radiologists can tweak the model predictions as required. The model has been deployed in practice. In conclusion, cartilage segmentation on DW-MRI images with modified U-Nets achieves accuracy that outperforms the human segmenter. Code is available at https://github.com/aakashrkaku/knee-cartilage-segmentation
DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation
Nov 14, 2019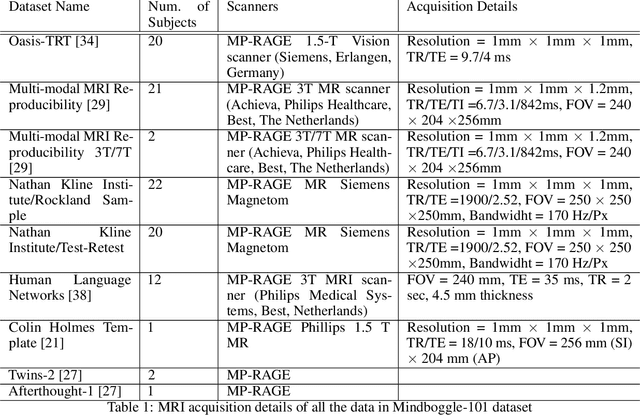
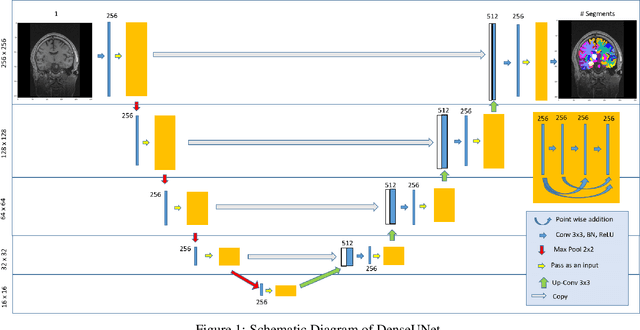
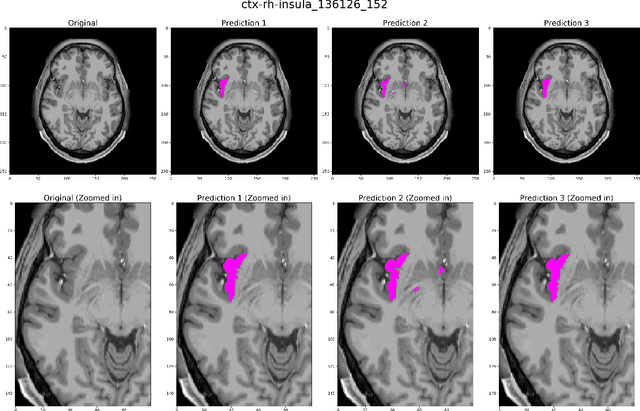
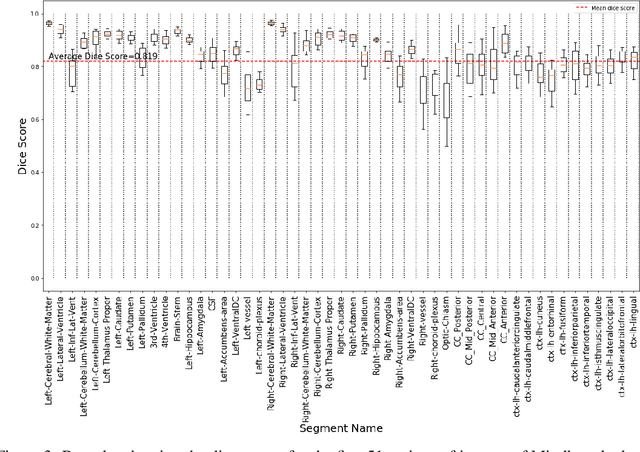
Abstract:Quantitative, volumetric analysis of Magnetic Resonance Imaging (MRI) is a fundamental way researchers study the brain in a host of neurological conditions including normal maturation and aging. Despite the availability of open-source brain segmentation software, widespread clinical adoption of volumetric analysis has been hindered due to processing times and reliance on manual corrections. Here, we extend the use of deep learning models from proof-of-concept, as previously reported, to present a comprehensive segmentation of cortical and deep gray matter brain structures matching the standard regions of aseg+aparc included in the commonly used open-source tool, Freesurfer. The work presented here provides a real-life, rapid deep learning-based brain segmentation tool to enable clinical translation as well as research application of quantitative brain segmentation. The advantages of the presented tool include short (~1 minute) processing time and improved segmentation quality. This is the first study to perform quick and accurate segmentation of 102 brain regions based on the surface-based protocol (DMK protocol), widely used by experts in the field. This is also the first work to include an expert reader study to assess the quality of the segmentation obtained using a deep-learning-based model. We show the superior performance of our deep-learning-based models over the traditional segmentation tool, Freesurfer. We refer to the proposed deep learning-based tool as DARTS (DenseUnet-based Automatic Rapid Tool for brain Segmentation). Our tool and trained models are available at https://github.com/NYUMedML/DARTS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge