Xiuyuan Wang
Convergence of the Min-Max Langevin Dynamics and Algorithm for Zero-Sum Games
Dec 29, 2024Abstract:We study zero-sum games in the space of probability distributions over the Euclidean space $\mathbb{R}^d$ with entropy regularization, in the setting when the interaction function between the players is smooth and strongly convex-concave. We prove an exponential convergence guarantee for the mean-field min-max Langevin dynamics to compute the equilibrium distribution of the zero-sum game. We also study the finite-particle approximation of the mean-field min-max Langevin dynamics, both in continuous and discrete times. We prove biased convergence guarantees for the continuous-time finite-particle min-max Langevin dynamics to the stationary mean-field equilibrium distribution with an explicit bias estimate which does not scale with the number of particles. We also prove biased convergence guarantees for the discrete-time finite-particle min-max Langevin algorithm to the stationary mean-field equilibrium distribution with an additional bias term which scales with the step size and the number of particles. This provides an explicit iteration complexity for the average particle along the finite-particle algorithm to approximately compute the equilibrium distribution of the zero-sum game.
A Symplectic Analysis of Alternating Mirror Descent
May 06, 2024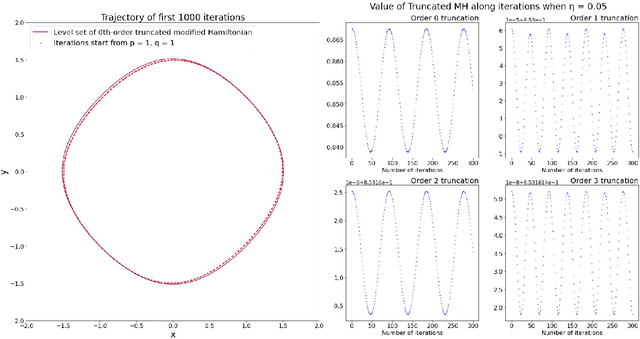
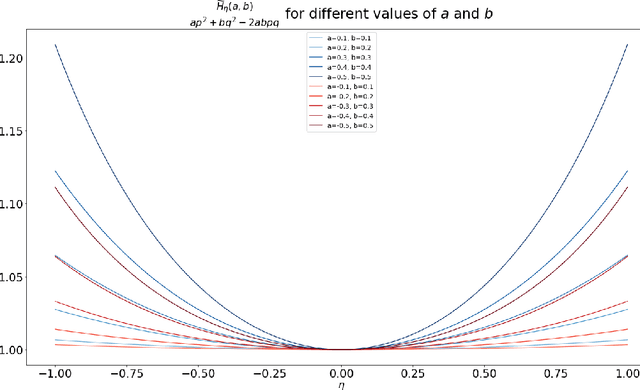
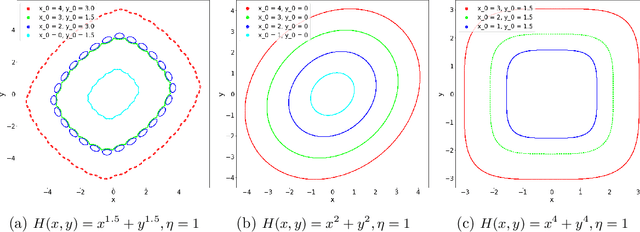
Abstract:Motivated by understanding the behavior of the Alternating Mirror Descent (AMD) algorithm for bilinear zero-sum games, we study the discretization of continuous-time Hamiltonian flow via the symplectic Euler method. We provide a framework for analysis using results from Hamiltonian dynamics, Lie algebra, and symplectic numerical integrators, with an emphasis on the existence and properties of a conserved quantity, the modified Hamiltonian (MH), for the symplectic Euler method. We compute the MH in closed-form when the original Hamiltonian is a quadratic function, and show that it generally differs from the other conserved quantity known previously in that case. We derive new error bounds on the MH when truncated at orders in the stepsize in terms of the number of iterations, $K$, and utilize this bound to show an improved $\mathcal{O}(K^{1/5})$ total regret bound and an $\mathcal{O}(K^{-4/5})$ duality gap of the average iterates for AMD. Finally, we propose a conjecture which, if true, would imply that the total regret for AMD goes as $\mathcal{O}\left(K^{\varepsilon}\right)$ and the duality gap of the average iterates as $\mathcal{O}\left(K^{-1+\varepsilon}\right)$ for any $\varepsilon>0$, and we can take $\varepsilon=0$ upon certain convergence conditions for the MH.
DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation
Nov 14, 2019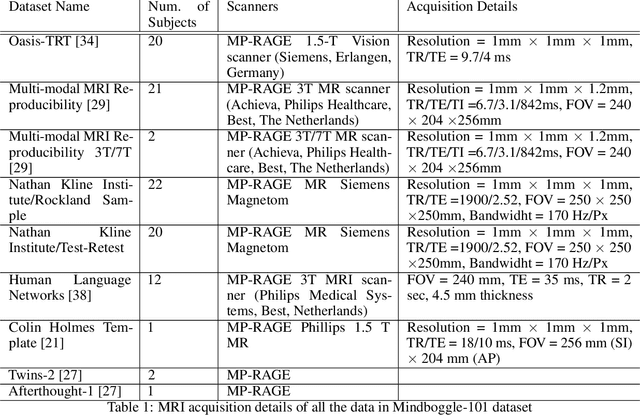
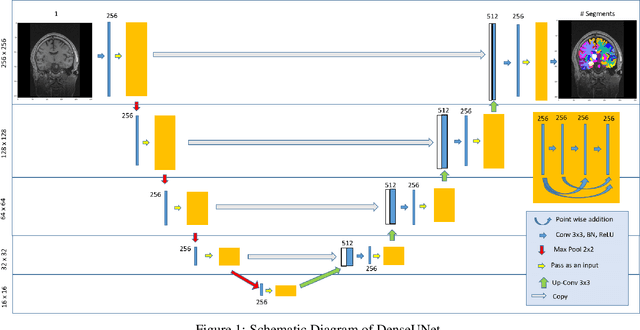
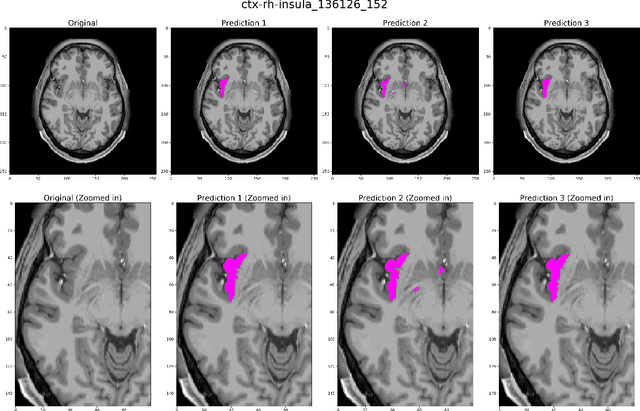
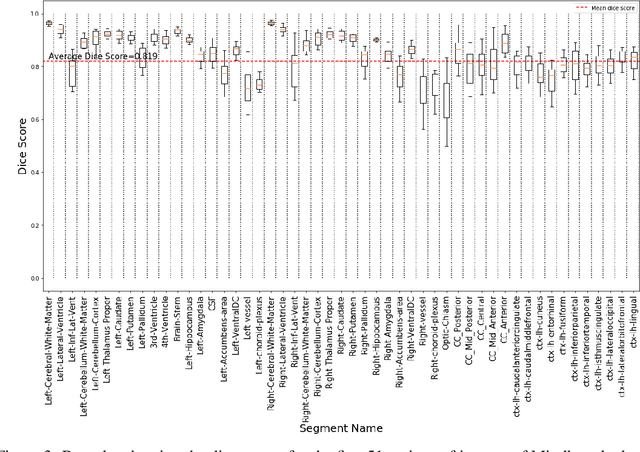
Abstract:Quantitative, volumetric analysis of Magnetic Resonance Imaging (MRI) is a fundamental way researchers study the brain in a host of neurological conditions including normal maturation and aging. Despite the availability of open-source brain segmentation software, widespread clinical adoption of volumetric analysis has been hindered due to processing times and reliance on manual corrections. Here, we extend the use of deep learning models from proof-of-concept, as previously reported, to present a comprehensive segmentation of cortical and deep gray matter brain structures matching the standard regions of aseg+aparc included in the commonly used open-source tool, Freesurfer. The work presented here provides a real-life, rapid deep learning-based brain segmentation tool to enable clinical translation as well as research application of quantitative brain segmentation. The advantages of the presented tool include short (~1 minute) processing time and improved segmentation quality. This is the first study to perform quick and accurate segmentation of 102 brain regions based on the surface-based protocol (DMK protocol), widely used by experts in the field. This is also the first work to include an expert reader study to assess the quality of the segmentation obtained using a deep-learning-based model. We show the superior performance of our deep-learning-based models over the traditional segmentation tool, Freesurfer. We refer to the proposed deep learning-based tool as DARTS (DenseUnet-based Automatic Rapid Tool for brain Segmentation). Our tool and trained models are available at https://github.com/NYUMedML/DARTS
Identification of relevant diffusion MRI metrics impacting cognitive functions using a novel feature selection method
Aug 10, 2019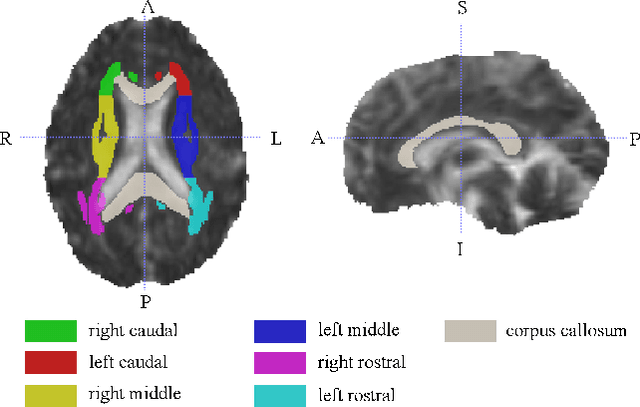
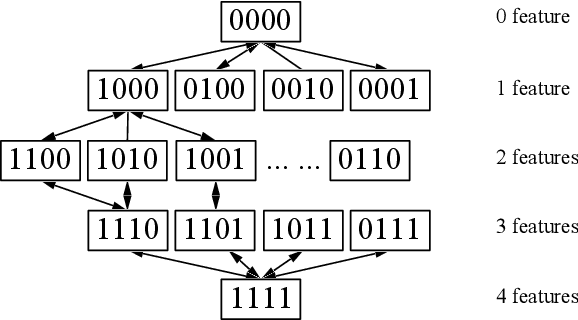
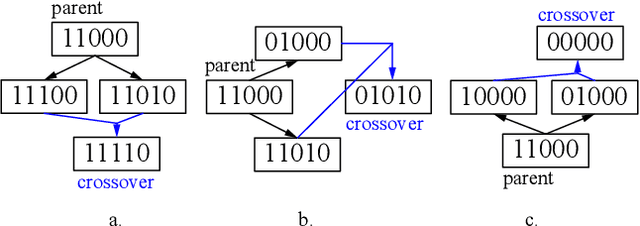
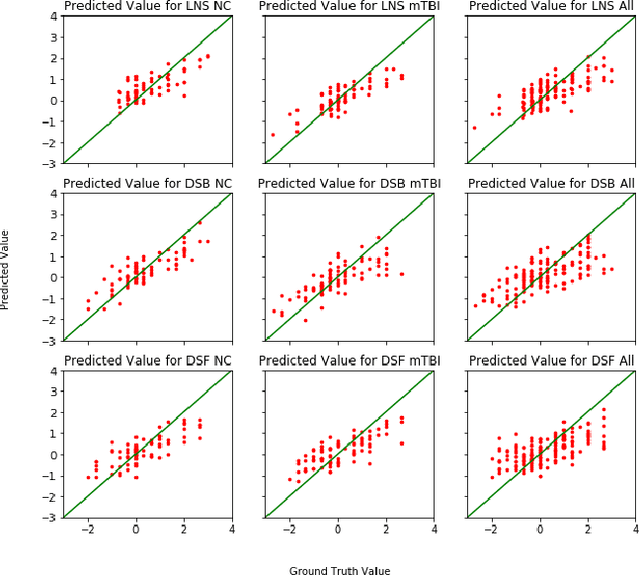
Abstract:Mild Traumatic Brain Injury (mTBI) is a significant public health problem. The most troubling symptoms after mTBI are cognitive complaints. Studies show measurable differences between patients with mTBI and healthy controls with respect to tissue microstructure using diffusion MRI. However, it remains unclear which diffusion measures are the most informative with regard to cognitive functions in both the healthy state as well as after injury. In this study, we use diffusion MRI to formulate a predictive model for performance on working memory based on the most relevant MRI features. The key challenge is to identify relevant features over a large feature space with high accuracy in an efficient manner. To tackle this challenge, we propose a novel improvement of the best first search approach with crossover operators inspired by genetic algorithm. Compared against other heuristic feature selection algorithms, the proposed method achieves significantly more accurate predictions and yields clinically interpretable selected features.
MTBI Identification From Diffusion MR Images Using Bag of Adversarial Visual Features
Jun 27, 2018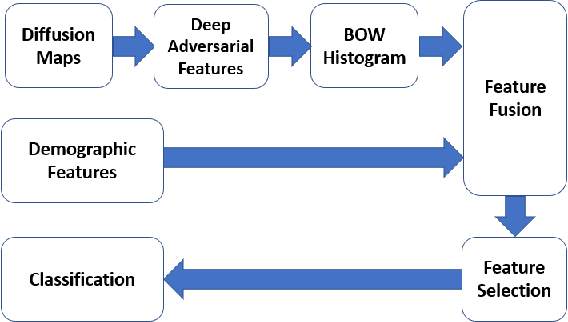
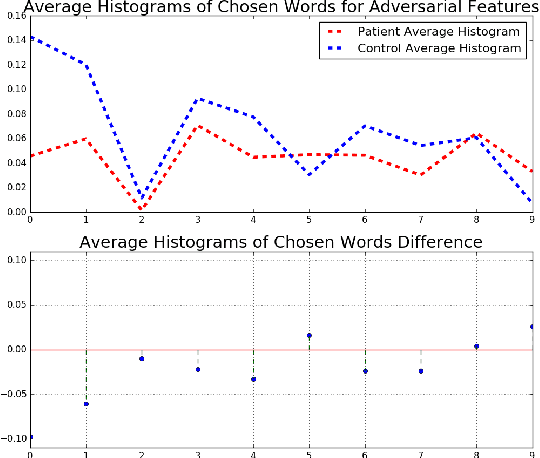
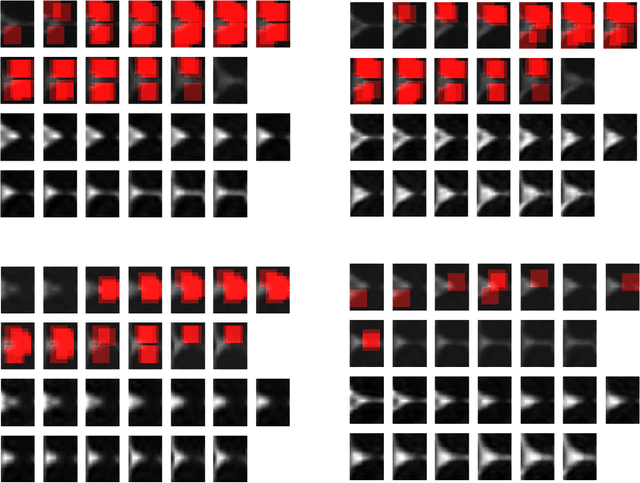
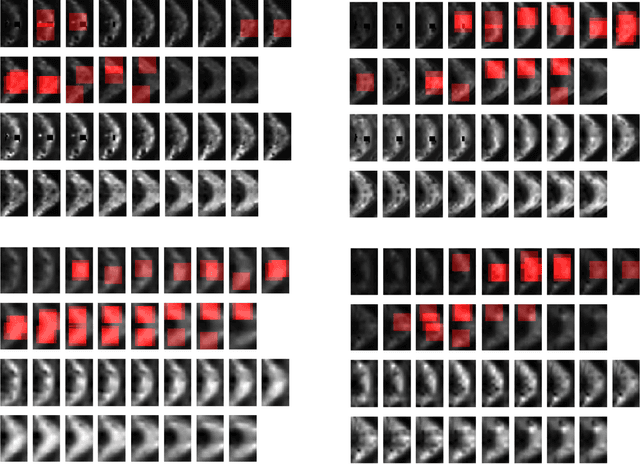
Abstract:In this work, we propose bag of adversarial features (BAF) for identifying mild traumatic brain injury (MTBI) patients from their diffusion magnetic resonance images (MRI) (obtained within one month of injury) by incorporating unsupervised feature learning techniques. MTBI is a growing public health problem with an estimated incidence of over 1.7 million people annually in US. Diagnosis is based on clinical history and symptoms, and accurate, concrete measures of injury are lacking. Unlike most of previous works, which use hand-crafted features extracted from different parts of brain for MTBI classification, we employ feature learning algorithms to learn more discriminative representation for this task. A major challenge in this field thus far is the relatively small number of subjects available for training. This makes it difficult to use an end-to-end convolutional neural network to directly classify a subject from MR images. To overcome this challenge, we first apply an adversarial auto-encoder (with convolutional structure) to learn patch-level features, from overlapping image patches extracted from different brain regions. We then aggregate these features through a bag-of-word approach. We perform an extensive experimental study on a dataset of 227 subjects (including 109 MTBI patients, and 118 age and sex matched healthy controls), and compare the bag-of-deep-features with several previous approaches. Our experimental results show that the BAF significantly outperforms earlier works relying on the mean values of MR metrics in selected brain regions.
A Deep Unsupervised Learning Approach Toward MTBI Identification Using Diffusion MRI
Apr 11, 2018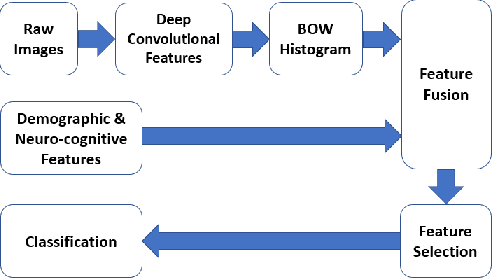
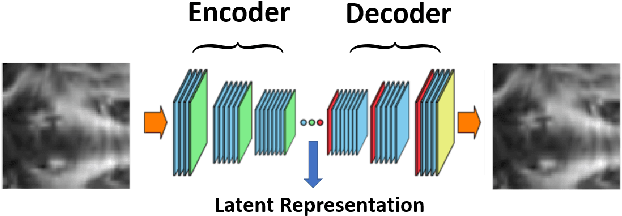
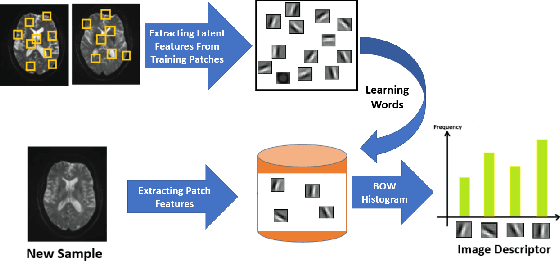
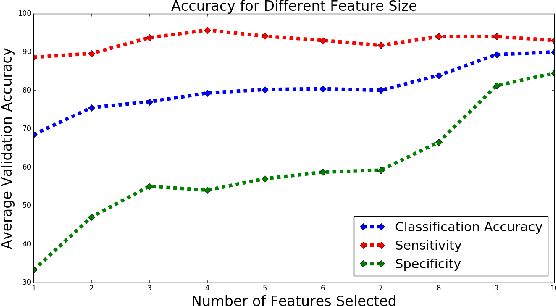
Abstract:Mild traumatic brain injury is a growing public health problem with an estimated incidence of over 1.7 million people annually in US. Diagnosis is based on clinical history and symptoms, and accurate, concrete measures of injury are lacking. This work aims to directly use diffusion MR images obtained within one month of trauma to detect injury, by incorporating deep learning techniques. To overcome the challenge due to limited training data, we describe each brain region using the bag of word representation, which specifies the distribution of representative patch patterns. We apply a convolutional auto-encoder to learn the patch-level features, from overlapping image patches extracted from the MR images, to learn features from diffusion MR images of brain using an unsupervised approach. Our experimental results show that the bag of word representation using patch level features learnt by the auto encoder provides similar performance as that using the raw patch patterns, both significantly outperform earlier work relying on the mean values of MR metrics in selected brain regions.
Identifying Mild Traumatic Brain Injury Patients From MR Images Using Bag of Visual Words
Feb 14, 2018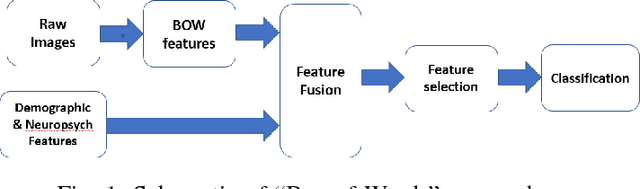
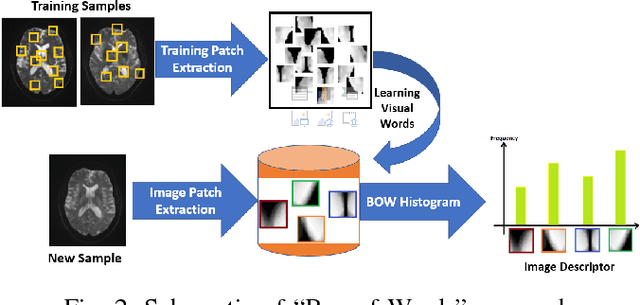
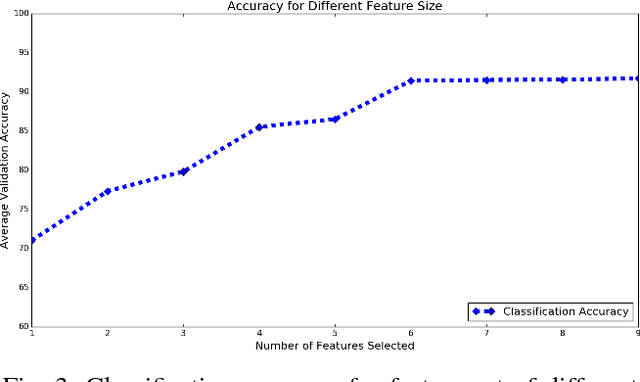
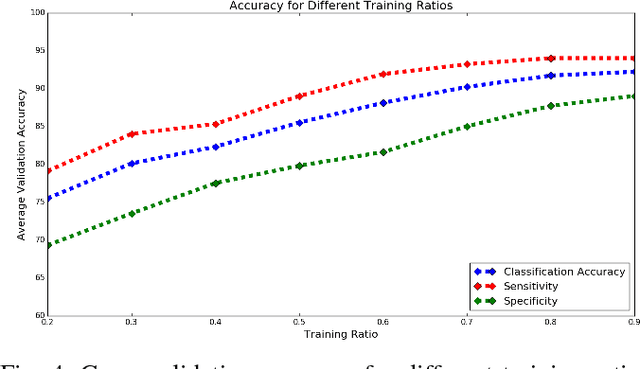
Abstract:Mild traumatic brain injury (mTBI) is a growing public health problem with an estimated incidence of one million people annually in US. Neurocognitive tests are used to both assess the patient condition and to monitor the patient progress. This work aims to directly use MR images taken shortly after injury to detect whether a patient suffers from mTBI, by incorporating machine learning and computer vision techniques to learn features suitable discriminating between mTBI and normal patients. We focus on 3 regions in brain, and extract multiple patches from them, and use bag-of-visual-word technique to represent each subject as a histogram of representative patterns derived from patches from all training subjects. After extracting the features, we use greedy forward feature selection, to choose a subset of features which achieves highest accuracy. We show through experimental studies that BoW features perform better than the simple mean value features which were used previously.
A Machine Learning Approach For Identifying Patients with Mild Traumatic Brain Injury Using Diffusion MRI Modeling
Aug 27, 2017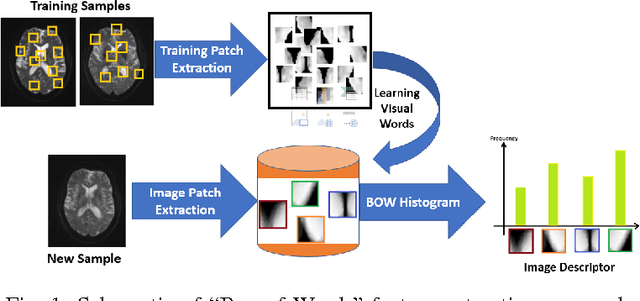
Abstract:While diffusion MRI has been extremely promising in the study of MTBI, identifying patients with recent MTBI remains a challenge. The literature is mixed with regard to localizing injury in these patients, however, gray matter such as the thalamus and white matter including the corpus callosum and frontal deep white matter have been repeatedly implicated as areas at high risk for injury. The purpose of this study is to develop a machine learning framework to classify MTBI patients and controls using features derived from multi-shell diffusion MRI in the thalamus, frontal white matter and corpus callosum.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge