Yvonne W Lui
FastMRI Prostate: A Publicly Available, Biparametric MRI Dataset to Advance Machine Learning for Prostate Cancer Imaging
Apr 18, 2023Abstract:The fastMRI brain and knee dataset has enabled significant advances in exploring reconstruction methods for improving speed and image quality for Magnetic Resonance Imaging (MRI) via novel, clinically relevant reconstruction approaches. In this study, we describe the April 2023 expansion of the fastMRI dataset to include biparametric prostate MRI data acquired on a clinical population. The dataset consists of raw k-space and reconstructed images for T2-weighted and diffusion-weighted sequences along with slice-level labels that indicate the presence and grade of prostate cancer. As has been the case with fastMRI, increasing accessibility to raw prostate MRI data will further facilitate research in MR image reconstruction and evaluation with the larger goal of improving the utility of MRI for prostate cancer detection and evaluation. The dataset is available at https://fastmri.med.nyu.edu.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Identification of relevant diffusion MRI metrics impacting cognitive functions using a novel feature selection method
Aug 10, 2019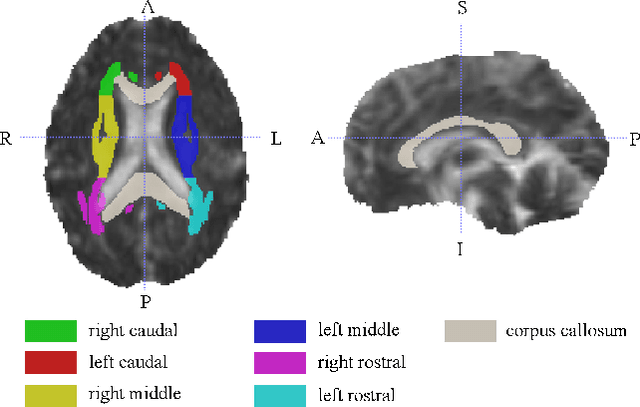
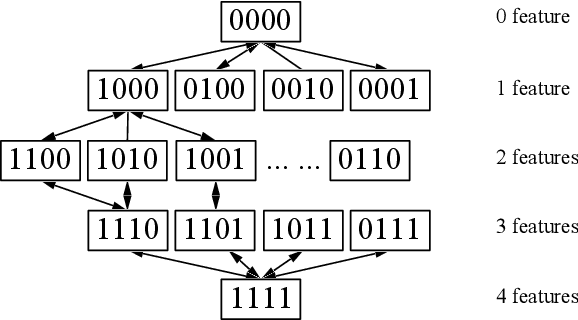
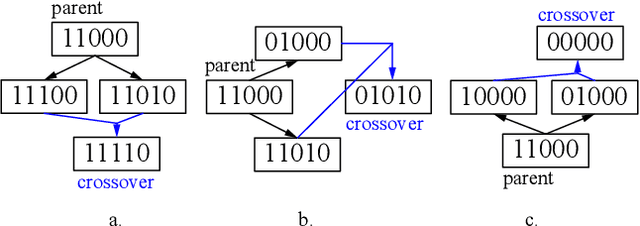
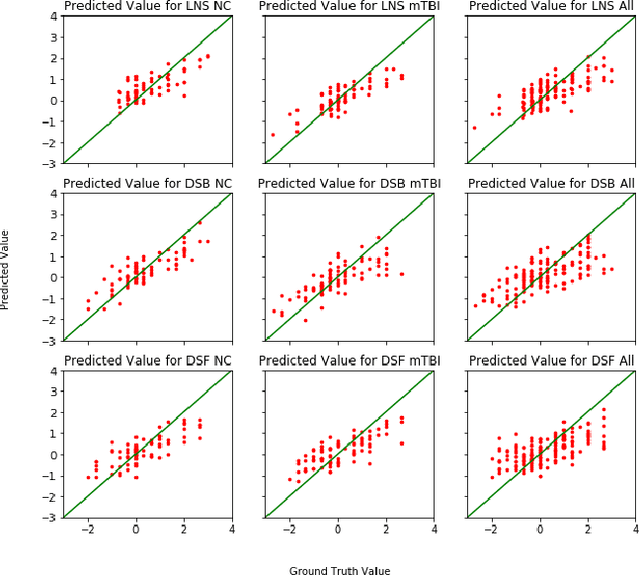
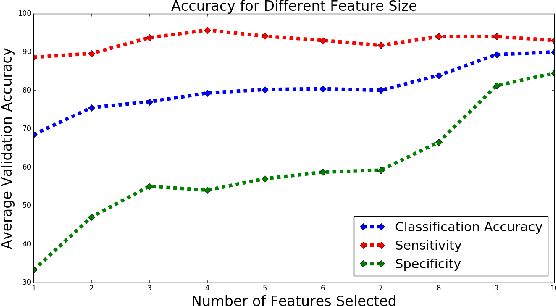
Abstract:Mild Traumatic Brain Injury (mTBI) is a significant public health problem. The most troubling symptoms after mTBI are cognitive complaints. Studies show measurable differences between patients with mTBI and healthy controls with respect to tissue microstructure using diffusion MRI. However, it remains unclear which diffusion measures are the most informative with regard to cognitive functions in both the healthy state as well as after injury. In this study, we use diffusion MRI to formulate a predictive model for performance on working memory based on the most relevant MRI features. The key challenge is to identify relevant features over a large feature space with high accuracy in an efficient manner. To tackle this challenge, we propose a novel improvement of the best first search approach with crossover operators inspired by genetic algorithm. Compared against other heuristic feature selection algorithms, the proposed method achieves significantly more accurate predictions and yields clinically interpretable selected features.
A Deep Unsupervised Learning Approach Toward MTBI Identification Using Diffusion MRI
Apr 11, 2018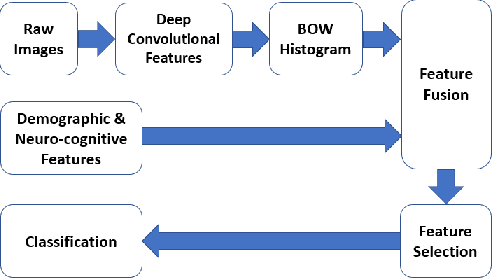
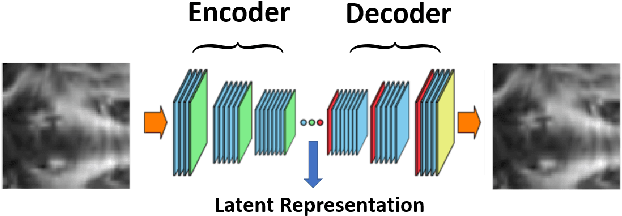
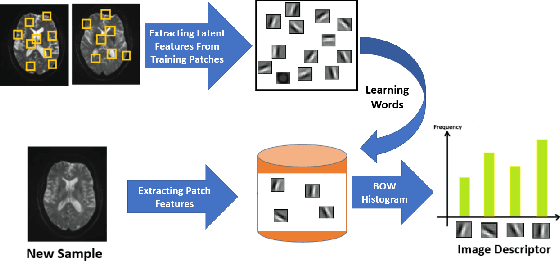
Abstract:Mild traumatic brain injury is a growing public health problem with an estimated incidence of over 1.7 million people annually in US. Diagnosis is based on clinical history and symptoms, and accurate, concrete measures of injury are lacking. This work aims to directly use diffusion MR images obtained within one month of trauma to detect injury, by incorporating deep learning techniques. To overcome the challenge due to limited training data, we describe each brain region using the bag of word representation, which specifies the distribution of representative patch patterns. We apply a convolutional auto-encoder to learn the patch-level features, from overlapping image patches extracted from the MR images, to learn features from diffusion MR images of brain using an unsupervised approach. Our experimental results show that the bag of word representation using patch level features learnt by the auto encoder provides similar performance as that using the raw patch patterns, both significantly outperform earlier work relying on the mean values of MR metrics in selected brain regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge