Radhika Tibrewala
fastMRI Breast: A publicly available radial k-space dataset of breast dynamic contrast-enhanced MRI
Jun 07, 2024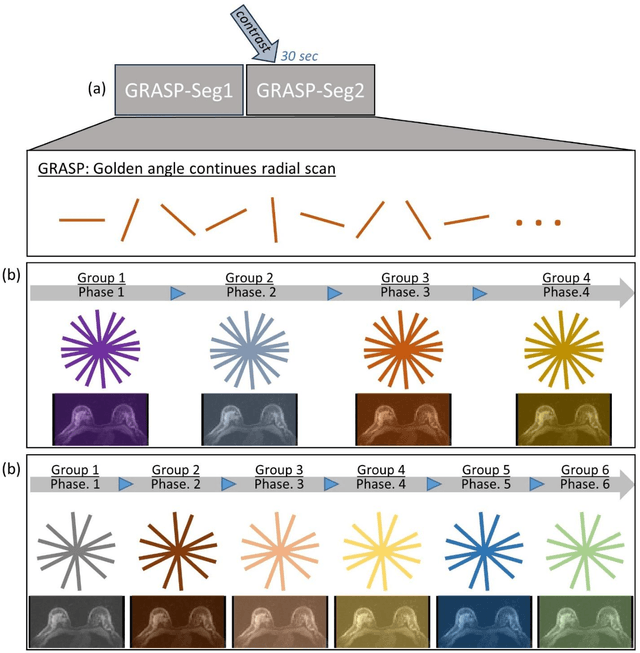
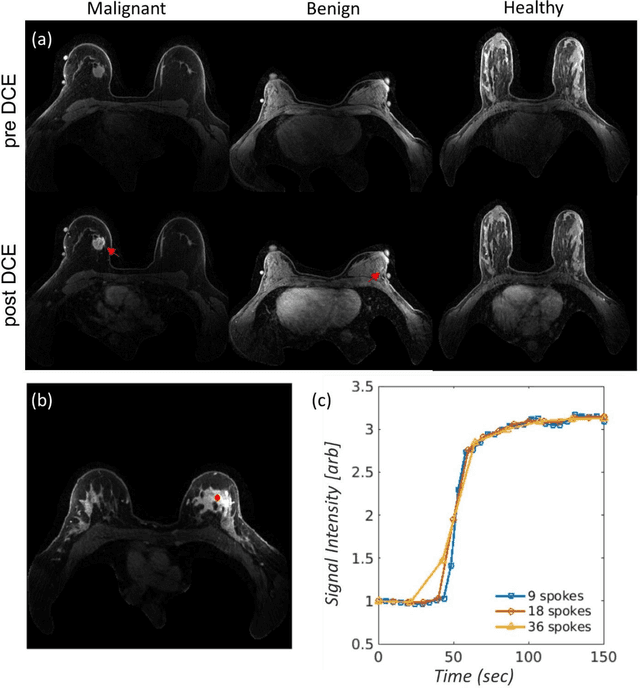
Abstract:This data curation work introduces the first large-scale dataset of radial k-space and DICOM data for breast DCE-MRI acquired in diagnostic breast MRI exams. Our dataset includes case-level labels indicating patient age, menopause status, lesion status (negative, benign, and malignant), and lesion type for each case. The public availability of this dataset and accompanying reconstruction code will support research and development of fast and quantitative breast image reconstruction and machine learning methods.
FastMRI Prostate: A Publicly Available, Biparametric MRI Dataset to Advance Machine Learning for Prostate Cancer Imaging
Apr 18, 2023Abstract:The fastMRI brain and knee dataset has enabled significant advances in exploring reconstruction methods for improving speed and image quality for Magnetic Resonance Imaging (MRI) via novel, clinically relevant reconstruction approaches. In this study, we describe the April 2023 expansion of the fastMRI dataset to include biparametric prostate MRI data acquired on a clinical population. The dataset consists of raw k-space and reconstructed images for T2-weighted and diffusion-weighted sequences along with slice-level labels that indicate the presence and grade of prostate cancer. As has been the case with fastMRI, increasing accessibility to raw prostate MRI data will further facilitate research in MR image reconstruction and evaluation with the larger goal of improving the utility of MRI for prostate cancer detection and evaluation. The dataset is available at https://fastmri.med.nyu.edu.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020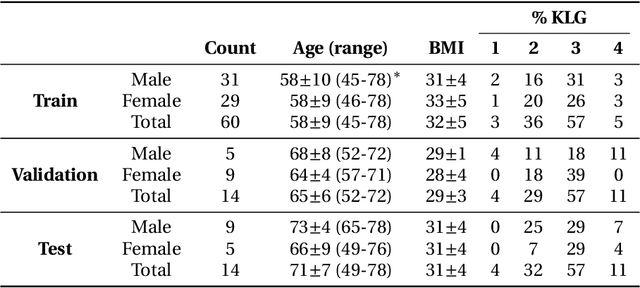
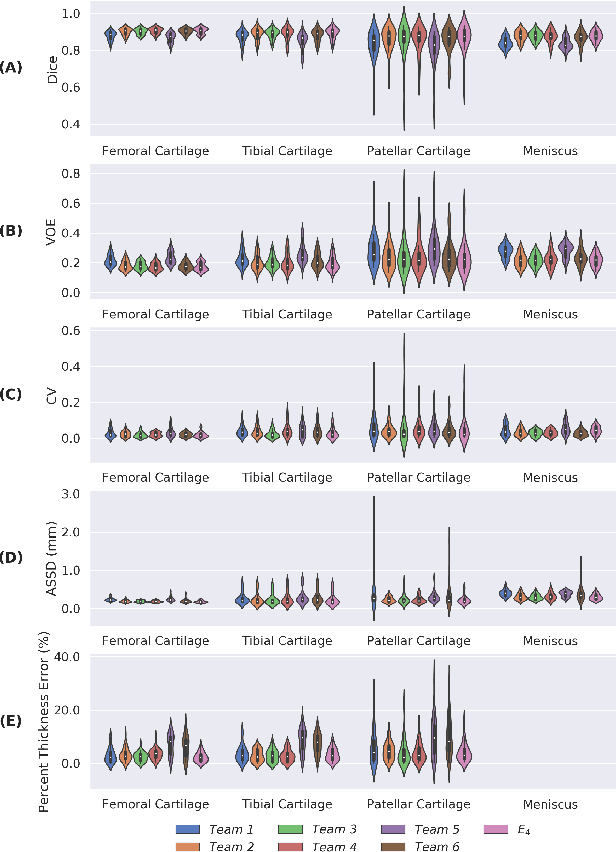
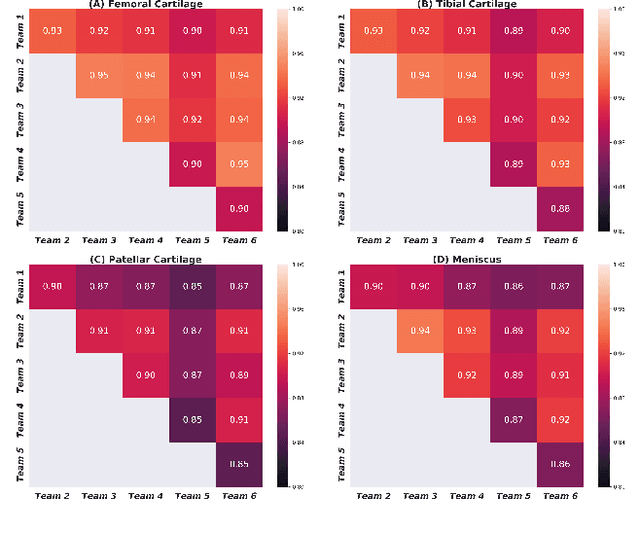
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Hierarchical Severity Staging of Anterior Cruciate Ligament Injuries using Deep Learning with MRI Images
Apr 13, 2020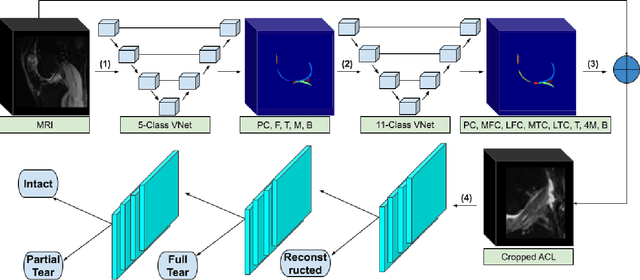
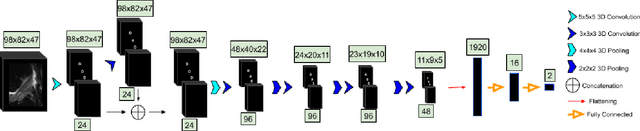
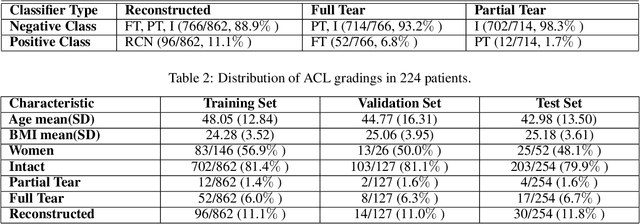
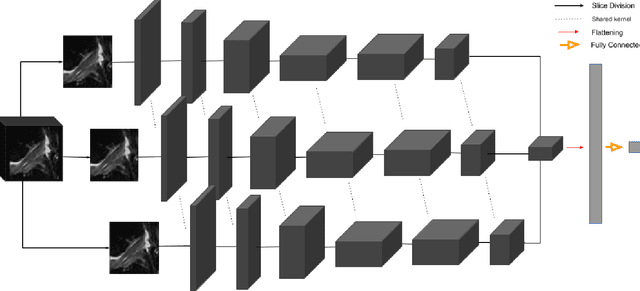
Abstract:Purpose: To evaluate the diagnostic utility of two convolutional neural networks (CNNs) for severity staging of anterior cruciate ligament (ACL) injuries. Materials and Methods: This retrospective analysis was conducted on 1243 knee MR images (1008 intact, 18 partially torn, 77 fully torn, and 140 reconstructed ACLs) from 224 patients (age 47 +/- 14 years, 54% women) acquired between 2011 and 2014. The radiologists used a modified scoring metric. To classify ACL injuries with deep learning, two types of CNNs were used, one with three-dimensional (3D) and the other with two-dimensional (2D) convolutional kernels. Performance metrics included sensitivity, specificity, weighted Cohen's kappa, and overall accuracy, followed by McNemar's test to compare the CNNs performance. Results: The overall accuracy and weighted Cohen's kappa reported for ACL injury classification were higher using the 2D CNN (accuracy: 92% (233/254) and kappa: 0.83) than the 3D CNN (accuracy: 89% (225/254) and kappa: 0.83) (P = .27). The 2D CNN and 3D CNN performed similarly in classifying intact ACLs (2D CNN: 93% (188/203) sensitivity and 90% (46/51) specificity; 3D CNN: 89% (180/203) sensitivity and 88% (45/51) specificity). Classification of full tears by both networks were also comparable (2D CNN: 82% (14/17) sensitivity and 94% (222/237) specificity; 3D CNN: 76% (13/17) sensitivity and 100% (236/237) specificity). The 2D CNN classified all reconstructed ACLs correctly. Conclusion: 2D and 3D CNNs applied to ACL lesion classification had high sensitivity and specificity, suggesting that these networks could be used to help grade ACL injuries by non-experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge