Florian Knoll
Motion-Guided Deep Image Prior for Cardiac MRI
Dec 05, 2024



Abstract:Cardiovascular magnetic resonance imaging is a powerful diagnostic tool for assessing cardiac structure and function. Traditional breath-held imaging protocols, however, pose challenges for patients with arrhythmias or limited breath-holding capacity. We introduce Motion-Guided Deep Image prior (M-DIP), a novel unsupervised reconstruction framework for accelerated real-time cardiac MRI. M-DIP employs a spatial dictionary to synthesize a time-dependent template image, which is further refined using time-dependent deformation fields that model cardiac and respiratory motion. Unlike prior DIP-based methods, M-DIP simultaneously captures physiological motion and frame-to-frame content variations, making it applicable to a wide range of dynamic applications. We validate M-DIP using simulated MRXCAT cine phantom data as well as free-breathing real-time cine and single-shot late gadolinium enhancement data from clinical patients. Comparative analyses against state-of-the-art supervised and unsupervised approaches demonstrate M-DIP's performance and versatility. M-DIP achieved better image quality metrics on phantom data, as well as higher reader scores for in-vivo patient data.
Domain Influence in MRI Medical Image Segmentation: spatial versus k-space inputs
Jul 01, 2024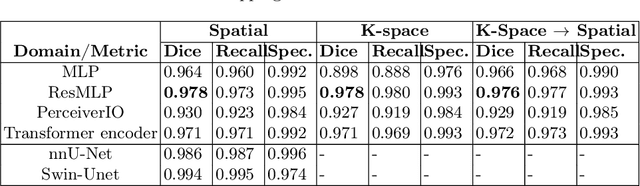


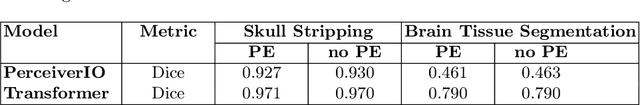
Abstract:Transformer-based networks applied to image patches have achieved cutting-edge performance in many vision tasks. However, lacking the built-in bias of convolutional neural networks (CNN) for local image statistics, they require large datasets and modifications to capture relationships between patches, especially in segmentation tasks. Images in the frequency domain might be more suitable for the attention mechanism, as local features are represented globally. By transforming images into the frequency domain, local features are represented globally. Due to MRI data acquisition properties, these images are particularly suitable. This work investigates how the image domain (spatial or k-space) affects segmentation results of deep learning (DL) models, focusing on attention-based networks and other non-convolutional models based on MLPs. We also examine the necessity of additional positional encoding for Transformer-based networks when input images are in the frequency domain. For evaluation, we pose a skull stripping task and a brain tissue segmentation task. The attention-based models used are PerceiverIO and a vanilla Transformer encoder. To compare with non-attention-based models, an MLP and ResMLP are also trained and tested. Results are compared with the Swin-Unet, the state-of-the-art medical image segmentation model. Experimental results show that using k-space for the input domain can significantly improve segmentation results. Also, additional positional encoding does not seem beneficial for attention-based networks if the input is in the frequency domain. Although none of the models matched the Swin-Unet's performance, the less complex models showed promising improvements with a different domain choice.
fastMRI Breast: A publicly available radial k-space dataset of breast dynamic contrast-enhanced MRI
Jun 07, 2024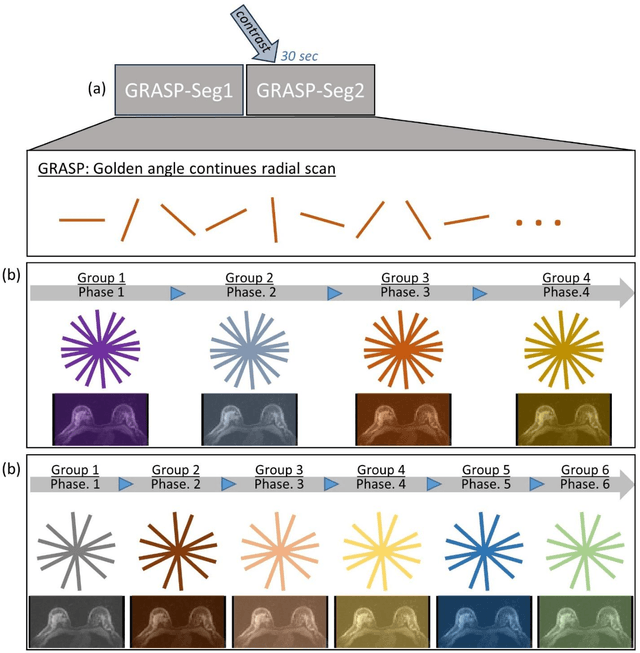
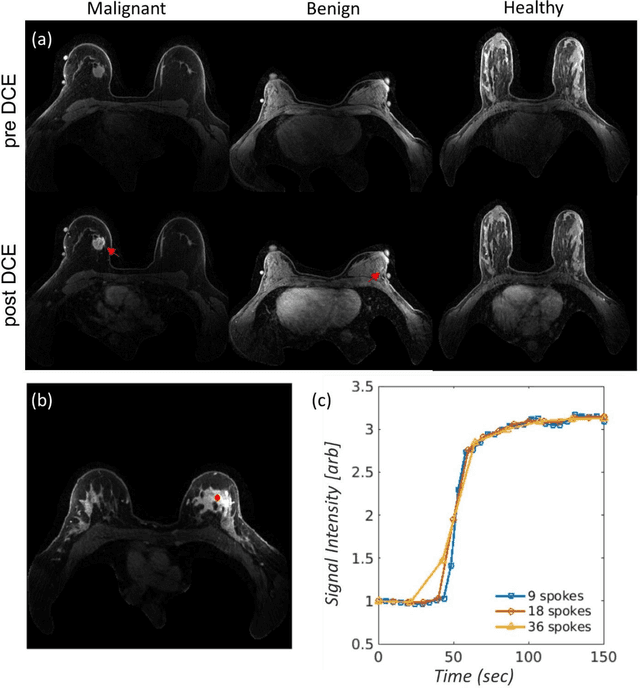
Abstract:This data curation work introduces the first large-scale dataset of radial k-space and DICOM data for breast DCE-MRI acquired in diagnostic breast MRI exams. Our dataset includes case-level labels indicating patient age, menopause status, lesion status (negative, benign, and malignant), and lesion type for each case. The public availability of this dataset and accompanying reconstruction code will support research and development of fast and quantitative breast image reconstruction and machine learning methods.
Accelerated MR Cholangiopancreatography with Deep Learning-based Reconstruction
May 06, 2024Abstract:This study accelerates MR cholangiopancreatography (MRCP) acquisitions using deep learning-based (DL) reconstruction at 3T and 0.55T. Thirty healthy volunteers underwent conventional two-fold MRCP scans at field strengths of 3T or 0.55T. We trained a variational network (VN) using retrospectively six-fold undersampled data obtained at 3T. We then evaluated our method against standard techniques such as parallel imaging (PI) and compressed sensing (CS), focusing on peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) as metrics. Furthermore, considering acquiring fully-sampled MRCP is impractical, we added a self-supervised DL reconstruction (SSDU) to the evaluating group. We also tested our method in a prospective accelerated scenario to reflect real-world clinical applications and evaluated its adaptability to MRCP at 0.55T. Our method demonstrated a remarkable reduction of average acquisition time from 599/542 to 255/180 seconds for MRCP at 3T/0.55T. In both retrospective and prospective undersampling scenarios, the PSNR and SSIM of VN were higher than those of PI, CS, and SSDU. At the same time, VN preserved the image quality of undersampled data, i.e., sharpness and the visibility of hepatobiliary ducts. In addition, VN also produced high quality reconstructions at 0.55T resulting in the highest PSNR and SSIM. In summary, VN trained for highly accelerated MRCP allows to reduce the acquisition time by a factor of 2.4/3.0 at 3T/0.55T while maintaining the image quality of the conventional acquisition.
Stable deep MRI reconstruction using Generative Priors
Oct 25, 2022



Abstract:Data-driven approaches recently achieved remarkable success in medical image reconstruction, but integration into clinical routine remains challenging due to a lack of generalizability and interpretability. Existing approaches usually require high-quality data-image pairs for training, but such data is not easily available for any imaging protocol and the reconstruction quality can quickly degrade even if only minor changes are made to the protocol. In addition, data-driven methods may create artificial features that can influence the clinicians decision-making. This is unacceptable if the clinician is unaware of the uncertainty associated with the reconstruction. In this paper, we address these challenges in a unified framework based on generative image priors. We propose a novel deep neural network based regularizer which is trained in an unsupervised setting on reference images without requiring any data-image pairs. After training, the regularizer can be used as part of a classical variational approach in combination with any acquisition protocols and shows stable behavior even if the test data deviates significantly from the training data. Furthermore, our probabilistic interpretation provides a distribution of reconstructions and hence allows uncertainty quantification. We demonstrate our approach on parallel magnetic resonance imaging, where results show competitive performance with SotA end-to-end deep learning methods, while preserving the flexibility of the acquisition protocol and allowing for uncertainty quantification.
Physics-Driven Deep Learning for Computational Magnetic Resonance Imaging
Mar 23, 2022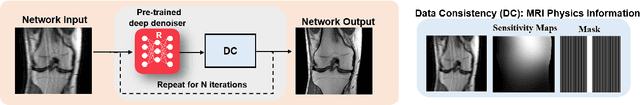
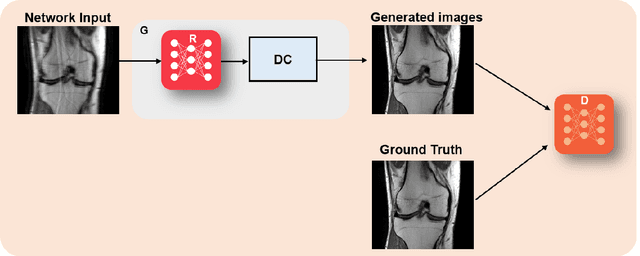
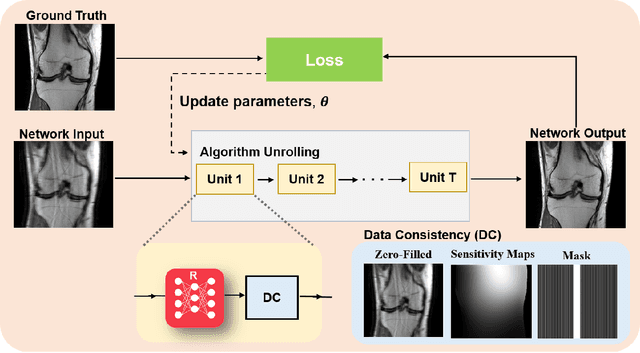
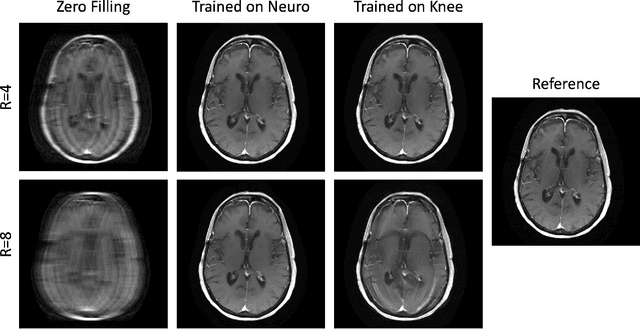
Abstract:Physics-driven deep learning methods have emerged as a powerful tool for computational magnetic resonance imaging (MRI) problems, pushing reconstruction performance to new limits. This article provides an overview of the recent developments in incorporating physics information into learning-based MRI reconstruction. We consider inverse problems with both linear and non-linear forward models for computational MRI, and review the classical approaches for solving these. We then focus on physics-driven deep learning approaches, covering physics-driven loss functions, plug-and-play methods, generative models, and unrolled networks. We highlight domain-specific challenges such as real- and complex-valued building blocks of neural networks, and translational applications in MRI with linear and non-linear forward models. Finally, we discuss common issues and open challenges, and draw connections to the importance of physics-driven learning when combined with other downstream tasks in the medical imaging pipeline.
Alternating Learning Approach for Variational Networks and Undersampling Pattern in Parallel MRI Applications
Oct 27, 2021
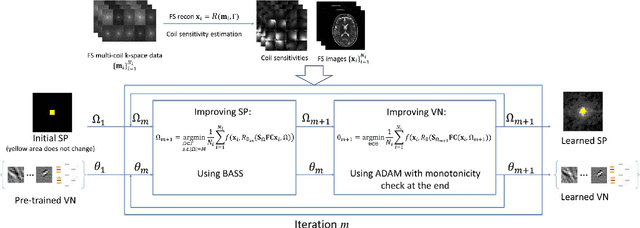


Abstract:Purpose: To propose an alternating learning approach to learn the sampling pattern (SP) and the parameters of variational networks (VN) in accelerated parallel magnetic resonance imaging (MRI). Methods: The approach alternates between improving the SP, using bias-accelerated subset selection, and improving parameters of the VN, using ADAM with monotonicity verification. The algorithm learns an effective pair: an SP that captures fewer k-space samples generating undersampling artifacts that are removed by the VN reconstruction. The proposed approach was tested for stability and convergence, considering different initial SPs. The quality of the VNs and SPs was compared against other approaches, including joint learning methods and VN learning with fixed variable density Poisson-disc SPs, using two different datasets and different acceleration factors (AF). Results: The root mean squared error (RMSE) improvements ranged from 14.9% to 51.2% considering AF from 2 to 20 in the tested brain and knee joint datasets when compared to the other approaches. The proposed approach has shown stable convergence, obtaining similar SPs with the same RMSE under different initial conditions. Conclusion: The proposed approach was stable and learned effective SPs with the corresponding VN parameters that produce images with better quality than other approaches, improving accelerated parallel MRI applications.
fastMRI+: Clinical Pathology Annotations for Knee and Brain Fully Sampled Multi-Coil MRI Data
Sep 14, 2021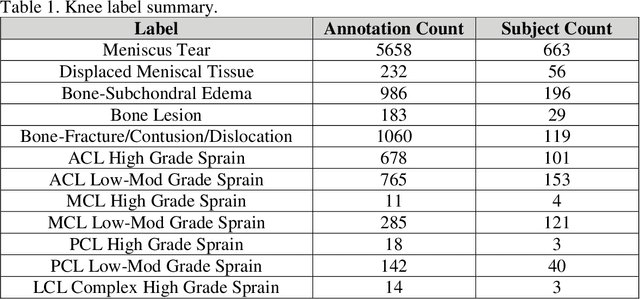
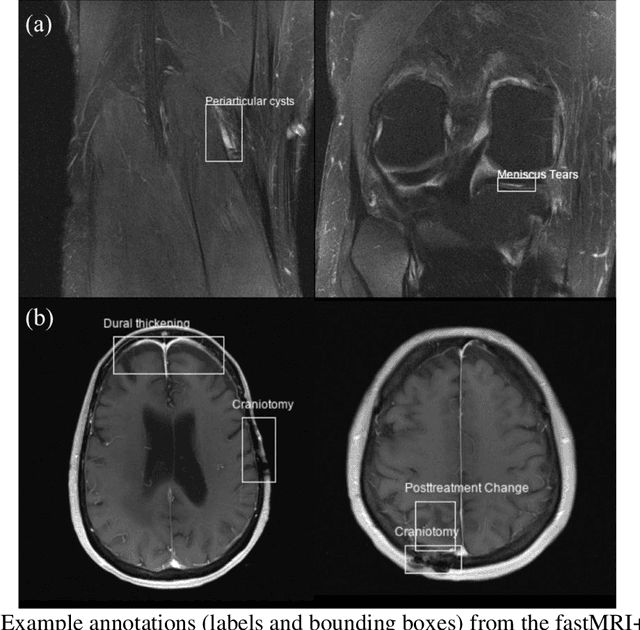
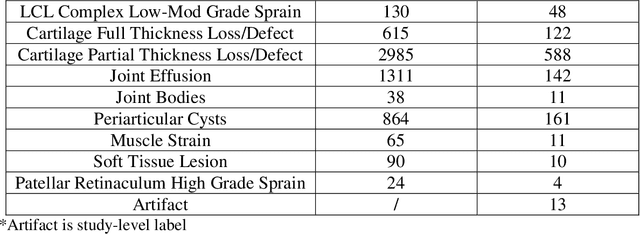
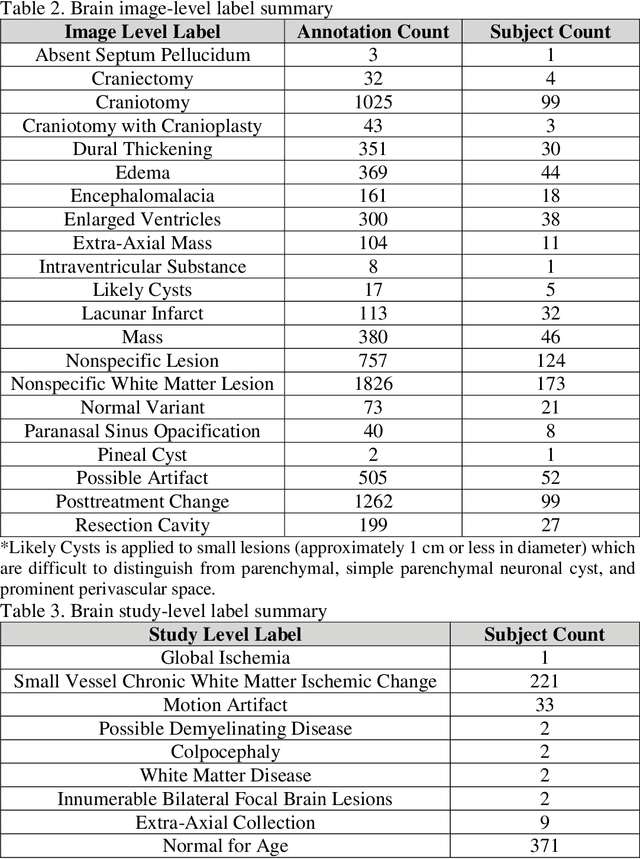
Abstract:Improving speed and image quality of Magnetic Resonance Imaging (MRI) via novel reconstruction approaches remains one of the highest impact applications for deep learning in medical imaging. The fastMRI dataset, unique in that it contains large volumes of raw MRI data, has enabled significant advances in accelerating MRI using deep learning-based reconstruction methods. While the impact of the fastMRI dataset on the field of medical imaging is unquestioned, the dataset currently lacks clinical expert pathology annotations, critical to addressing clinically relevant reconstruction frameworks and exploring important questions regarding rendering of specific pathology using such novel approaches. This work introduces fastMRI+, which consists of 16154 subspecialist expert bounding box annotations and 13 study-level labels for 22 different pathology categories on the fastMRI knee dataset, and 7570 subspecialist expert bounding box annotations and 643 study-level labels for 30 different pathology categories for the fastMRI brain dataset. The fastMRI+ dataset is open access and aims to support further research and advancement of medical imaging in MRI reconstruction and beyond.
Bayesian Uncertainty Estimation of Learned Variational MRI Reconstruction
Feb 12, 2021



Abstract:Recent deep learning approaches focus on improving quantitative scores of dedicated benchmarks, and therefore only reduce the observation-related (aleatoric) uncertainty. However, the model-immanent (epistemic) uncertainty is less frequently systematically analyzed. In this work, we introduce a Bayesian variational framework to quantify the epistemic uncertainty. To this end, we solve the linear inverse problem of undersampled MRI reconstruction in a variational setting. The associated energy functional is composed of a data fidelity term and the total deep variation (TDV) as a learned parametric regularizer. To estimate the epistemic uncertainty we draw the parameters of the TDV regularizer from a multivariate Gaussian distribution, whose mean and covariance matrix are learned in a stochastic optimal control problem. In several numerical experiments, we demonstrate that our approach yields competitive results for undersampled MRI reconstruction. Moreover, we can accurately quantify the pixelwise epistemic uncertainty, which can serve radiologists as an additional resource to visualize reconstruction reliability.
State-of-the-Art Machine Learning MRI Reconstruction in 2020: Results of the Second fastMRI Challenge
Dec 28, 2020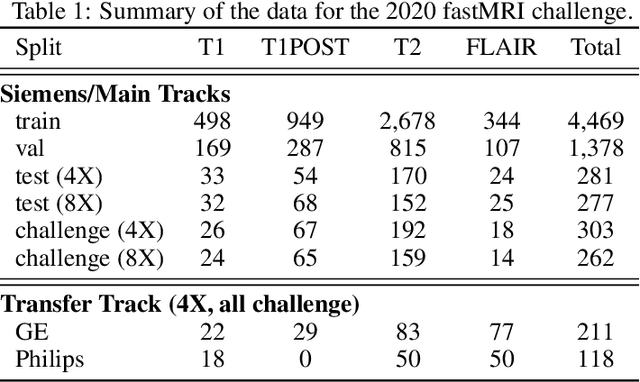
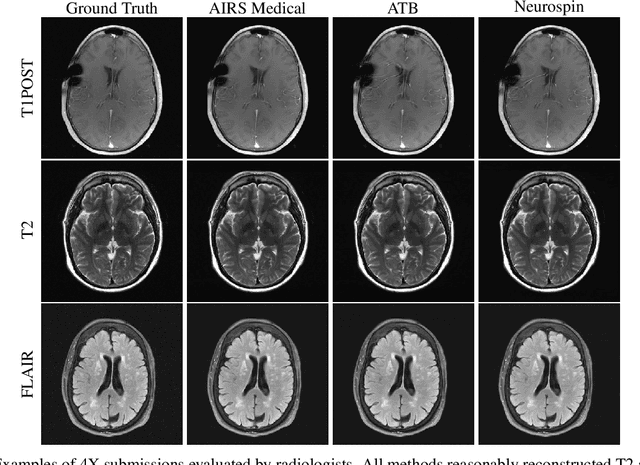
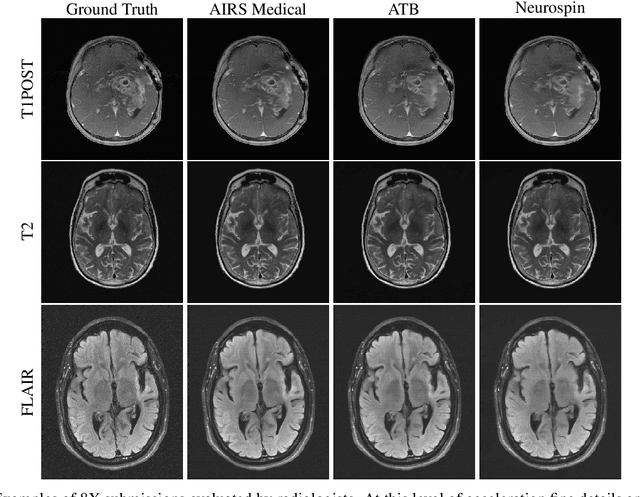
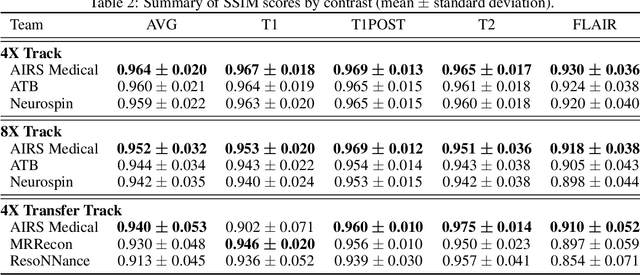
Abstract:Accelerating MRI scans is one of the principal outstanding problems in the MRI research community. Towards this goal, we hosted the second fastMRI competition targeted towards reconstructing MR images with subsampled k-space data. We provided participants with data from 7,299 clinical brain scans (de-identified via a HIPAA-compliant procedure by NYU Langone Health), holding back the fully-sampled data from 894 of these scans for challenge evaluation purposes. In contrast to the 2019 challenge, we focused our radiologist evaluations on pathological assessment in brain images. We also debuted a new Transfer track that required participants to submit models evaluated on MRI scanners from outside the training set. We received 19 submissions from eight different groups. Results showed one team scoring best in both SSIM scores and qualitative radiologist evaluations. We also performed analysis on alternative metrics to mitigate the effects of background noise and collected feedback from the participants to inform future challenges. Lastly, we identify common failure modes across the submissions, highlighting areas of need for future research in the MRI reconstruction community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge