Marcel Dominik Nickel
Accelerated MR Cholangiopancreatography with Deep Learning-based Reconstruction
May 06, 2024Abstract:This study accelerates MR cholangiopancreatography (MRCP) acquisitions using deep learning-based (DL) reconstruction at 3T and 0.55T. Thirty healthy volunteers underwent conventional two-fold MRCP scans at field strengths of 3T or 0.55T. We trained a variational network (VN) using retrospectively six-fold undersampled data obtained at 3T. We then evaluated our method against standard techniques such as parallel imaging (PI) and compressed sensing (CS), focusing on peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) as metrics. Furthermore, considering acquiring fully-sampled MRCP is impractical, we added a self-supervised DL reconstruction (SSDU) to the evaluating group. We also tested our method in a prospective accelerated scenario to reflect real-world clinical applications and evaluated its adaptability to MRCP at 0.55T. Our method demonstrated a remarkable reduction of average acquisition time from 599/542 to 255/180 seconds for MRCP at 3T/0.55T. In both retrospective and prospective undersampling scenarios, the PSNR and SSIM of VN were higher than those of PI, CS, and SSDU. At the same time, VN preserved the image quality of undersampled data, i.e., sharpness and the visibility of hepatobiliary ducts. In addition, VN also produced high quality reconstructions at 0.55T resulting in the highest PSNR and SSIM. In summary, VN trained for highly accelerated MRCP allows to reduce the acquisition time by a factor of 2.4/3.0 at 3T/0.55T while maintaining the image quality of the conventional acquisition.
Deep learning-guided weighted averaging for signal dropout compensation in diffusion-weighted imaging of the liver
Feb 20, 2022Abstract:Purpose: To develop an algorithm for the retrospective correction of signal dropout artifacts in abdominal diffusion-weighted imaging (DWI) resulting from cardiac motion. Methods: Given a set of image repetitions for a slice, a locally adaptive weighted averaging is proposed which aims to suppress the contribution of image regions affected by signal dropouts. Corresponding weight maps were estimated by a sliding-window algorithm which analyzed signal deviations from a patch-wise reference. In order to ensure the computation of a robust reference, repetitions were filtered by a classifier that was trained to detect images corrupted by signal dropouts. The proposed method, termed Deep Learning-guided Adaptive Weighted Averaging (DLAWA), was evaluated in terms of dropout suppression capability, bias reduction in the Apparent Diffusion Coefficient (ADC) and noise characteristics. Results: In the case of uniform averaging, motion-related dropouts caused signal attenuation and ADC overestimation in parts of the liver with the left lobe being affected particularly. Both effects could be substantially mitigated by DLAWA while preventing global penalties with respect to signal-to-noise ratio (SNR) due to local signal suppression. Performing evaluations on patient data, the capability to recover lesions concealed by signal dropouts was demonstrated as well. Further, DLAWA allowed for transparent control of the trade-off between SNR and signal dropout suppression by means of a few hyperparameters. Conclusion: This work presents an effective and flexible method for the local compensation of signal dropouts resulting from motion and pulsation. Since DLAWA follows a retrospective approach, no changes to the acquisition are required.
Robust partial Fourier reconstruction for diffusion-weighted imaging using a recurrent convolutional neural network
May 19, 2021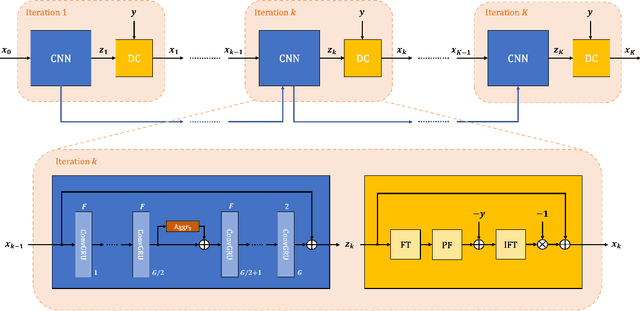
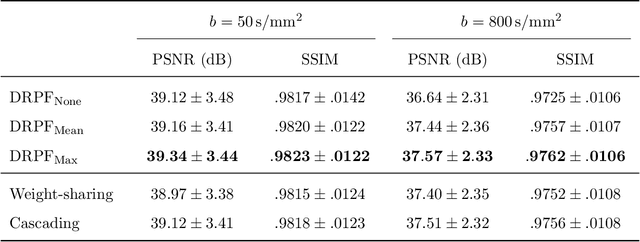
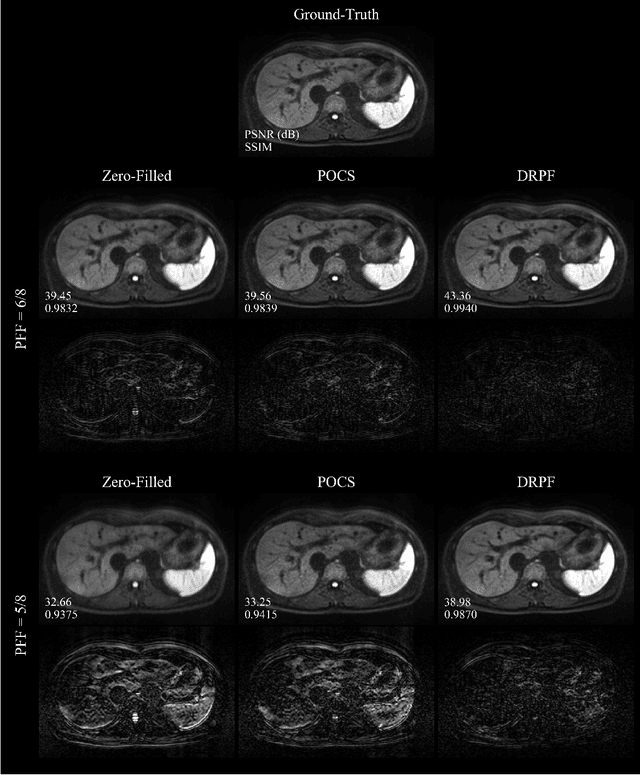
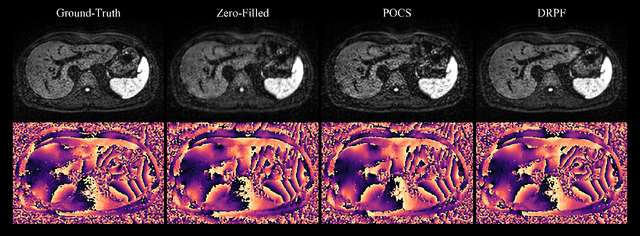
Abstract:Purpose: To develop an algorithm for robust partial Fourier (PF) reconstruction applicable to diffusion-weighted (DW) images with non-smooth phase variations. Methods: Based on an unrolled proximal splitting algorithm, a neural network architecture is derived which alternates between data consistency operations and regularization implemented by recurrent convolutions. In order to exploit correlations, multiple repetitions of the same slice are jointly reconstructed under consideration of permutation-equivariance. The proposed method is trained on DW liver data of 60 volunteers and evaluated on retrospectively and prospectively sub-sampled data of different anatomies and resolutions. In addition, the benefits of using a recurrent network over other unrolling strategies is investigated. Results: Conventional PF techniques can be significantly outperformed in terms of quantitative measures as well as perceptual image quality. The proposed method is able to generalize well to brain data with contrasts and resolution not present in the training set. The reduction in echo time (TE) associated with prospective PF-sampling enables DW imaging with higher signal. Also, the TE increase in acquisitions with higher resolution can be compensated for. It can be shown that unrolling by means of a recurrent network produced better results than using a weight-shared network or a cascade of networks. Conclusion: This work demonstrates that robust PF reconstruction of DW data is feasible even at strong PF factors in applications with severe phase variations. Since the proposed method does not rely on smoothness priors of the phase but uses learned recurrent convolutions instead, artifacts of conventional PF methods can be avoided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge