Mehmet Akçakaya
Deploying AI for Signal Processing education: Selected challenges and intriguing opportunities
Sep 10, 2025Abstract:Powerful artificial intelligence (AI) tools that have emerged in recent years -- including large language models, automated coding assistants, and advanced image and speech generation technologies -- are the result of monumental human achievements. These breakthroughs reflect mastery across multiple technical disciplines and the resolution of significant technological challenges. However, some of the most profound challenges may still lie ahead. These challenges are not purely technical but pertain to the fair and responsible use of AI in ways that genuinely improve the global human condition. This article explores one promising application aligned with that vision: the use of AI tools to facilitate and enhance education, with a specific focus on signal processing (SP). It presents two interrelated perspectives: identifying and addressing technical limitations, and applying AI tools in practice to improve educational experiences. Primers are provided on several core technical issues that arise when using AI in educational settings, including how to ensure fairness and inclusivity, handle hallucinated outputs, and achieve efficient use of resources. These and other considerations -- such as transparency, explainability, and trustworthiness -- are illustrated through the development of an immersive, structured, and reliable "smart textbook." The article serves as a resource for researchers and educators seeking to advance AI's role in engineering education.
Sparsity-Driven Parallel Imaging Consistency for Improved Self-Supervised MRI Reconstruction
May 30, 2025Abstract:Physics-driven deep learning (PD-DL) models have proven to be a powerful approach for improved reconstruction of rapid MRI scans. In order to train these models in scenarios where fully-sampled reference data is unavailable, self-supervised learning has gained prominence. However, its application at high acceleration rates frequently introduces artifacts, compromising image fidelity. To mitigate this shortcoming, we propose a novel way to train PD-DL networks via carefully-designed perturbations. In particular, we enhance the k-space masking idea of conventional self-supervised learning with a novel consistency term that assesses the model's ability to accurately predict the added perturbations in a sparse domain, leading to more reliable and artifact-free reconstructions. The results obtained from the fastMRI knee and brain datasets show that the proposed training strategy effectively reduces aliasing artifacts and mitigates noise amplification at high acceleration rates, outperforming state-of-the-art self-supervised methods both visually and quantitatively.
Deep Learning Assisted Outer Volume Removal for Highly-Accelerated Real-Time Dynamic MRI
May 01, 2025



Abstract:Real-time (RT) dynamic MRI plays a vital role in capturing rapid physiological processes, offering unique insights into organ motion and function. Among these applications, RT cine MRI is particularly important for functional assessment of the heart with high temporal resolution. RT imaging enables free-breathing, ungated imaging of cardiac motion, making it a crucial alternative for patients who cannot tolerate conventional breath-hold, ECG-gated acquisitions. However, achieving high acceleration rates in RT cine MRI is challenging due to aliasing artifacts from extra-cardiac tissues, particularly at high undersampling factors. In this study, we propose a novel outer volume removal (OVR) method to address this challenge by eliminating aliasing contributions from non-cardiac regions in a post-processing framework. Our approach estimates the outer volume signal for each timeframe using composite temporal images from time-interleaved undersampling patterns, which inherently contain pseudo-periodic ghosting artifacts. A deep learning (DL) model is trained to identify and remove these artifacts, producing a clean outer volume estimate that is subsequently subtracted from the corresponding k-space data. The final reconstruction is performed with a physics-driven DL (PD-DL) method trained using an OVR-specific loss function to restore high spatio-temporal resolution images. Experimental results show that the proposed method at high accelerations achieves image quality that is visually comparable to clinical baseline images, while outperforming conventional reconstruction techniques, both qualitatively and quantitatively. The proposed approach provides a practical and effective solution for artifact reduction in RT cine MRI without requiring acquisition modifications, offering a pathway to higher acceleration rates while preserving diagnostic quality.
Generative Model-Based Fusion for Improved Few-Shot Semantic Segmentation of Infrared Images
Dec 06, 2024
Abstract:Infrared (IR) imaging is commonly used in various scenarios, including autonomous driving, fire safety and defense applications. Thus, semantic segmentation of such images is of great interest. However, this task faces several challenges, including data scarcity, differing contrast and input channel number compared to natural images, and emergence of classes not represented in databases in certain scenarios, such as defense applications. Few-shot segmentation (FSS) provides a framework to overcome these issues by segmenting query images using a few labeled support samples. However, existing FSS models for IR images require paired visible RGB images, which is a major limitation since acquiring such paired data is difficult or impossible in some applications. In this work, we develop new strategies for FSS of IR images by using generative modeling and fusion techniques. To this end, we propose to synthesize auxiliary data to provide additional channel information to complement the limited contrast in the IR images, as well as IR data synthesis for data augmentation. Here, the former helps the FSS model to better capture the relationship between the support and query sets, while the latter addresses the issue of data scarcity. Finally, to further improve the former aspect, we propose a novel fusion ensemble module for integrating the two different modalities. Our methods are evaluated on different IR datasets, and improve upon the state-of-the-art (SOTA) FSS models.
Training Physics-Driven Deep Learning Reconstruction without Raw Data Access for Equitable Fast MRI
Nov 20, 2024
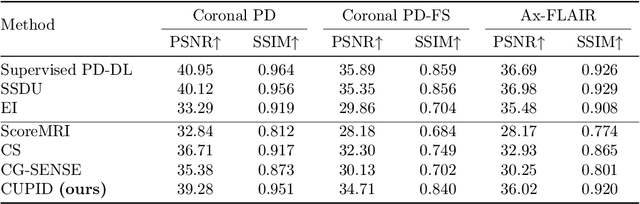

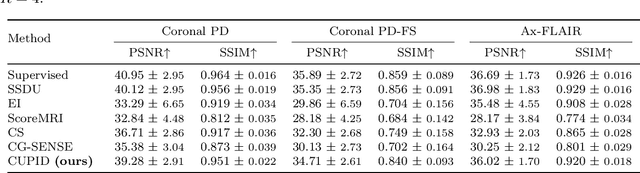
Abstract:Physics-driven deep learning (PD-DL) approaches have become popular for improved reconstruction of fast magnetic resonance imaging (MRI) scans. Even though PD-DL offers higher acceleration rates compared to existing clinical fast MRI techniques, their use has been limited outside specialized MRI centers. One impediment for their deployment is the difficulties with generalization to pathologies or population groups that are not well-represented in training sets. This has been noted in several studies, and fine-tuning on target populations to improve reconstruction has been suggested. However, current approaches for PD-DL training require access to raw k-space measurements, which is typically only available at specialized MRI centers that have research agreements for such data access. This is especially an issue for rural and underserved areas, where commercial MRI scanners only provide access to a final reconstructed image. To tackle these challenges, we propose Compressibility-inspired Unsupervised Learning via Parallel Imaging Fidelity (CUPID) for high-quality PD-DL training, using only routine clinical reconstructed images exported from an MRI scanner. CUPID evaluates the goodness of the output with a compressibility-based approach, while ensuring that the output stays consistent with the clinical parallel imaging reconstruction through well-designed perturbations. Our results show that CUPID achieves similar quality compared to well-established PD-DL training strategies that require raw k-space data access, while outperforming conventional compressed sensing (CS) and state-of-the-art generative methods. We also demonstrate its effectiveness in a zero-shot training setup for retrospectively and prospectively sub-sampled acquisitions, attesting to its minimal training burden.
Zero-Shot Adaptation for Approximate Posterior Sampling of Diffusion Models in Inverse Problems
Jul 16, 2024Abstract:Diffusion models have emerged as powerful generative techniques for solving inverse problems. Despite their success in a variety of inverse problems in imaging, these models require many steps to converge, leading to slow inference time. Recently, there has been a trend in diffusion models for employing sophisticated noise schedules that involve more frequent iterations of timesteps at lower noise levels, thereby improving image generation and convergence speed. However, application of these ideas for solving inverse problems with diffusion models remain challenging, as these noise schedules do not perform well when using empirical tuning for the forward model log-likelihood term weights. To tackle these challenges, we propose zero-shot approximate posterior sampling (ZAPS) that leverages connections to zero-shot physics-driven deep learning. ZAPS fixes the number of sampling steps, and uses zero-shot training with a physics-guided loss function to learn log-likelihood weights at each irregular timestep. We apply ZAPS to the recently proposed diffusion posterior sampling method as baseline, though ZAPS can also be used with other posterior sampling diffusion models. We further approximate the Hessian of the logarithm of the prior using a diagonalization approach with learnable diagonal entries for computational efficiency. These parameters are optimized over a fixed number of epochs with a given computational budget. Our results for various noisy inverse problems, including Gaussian and motion deblurring, inpainting, and super-resolution show that ZAPS reduces inference time, provides robustness to irregular noise schedules and improves reconstruction quality. Code is available at https://github.com/ualcalar17/ZAPS
Non-Cartesian Self-Supervised Physics-Driven Deep Learning Reconstruction for Highly-Accelerated Multi-Echo Spiral fMRI
Dec 09, 2023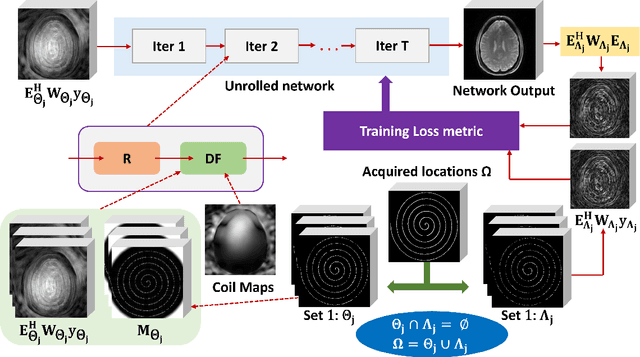
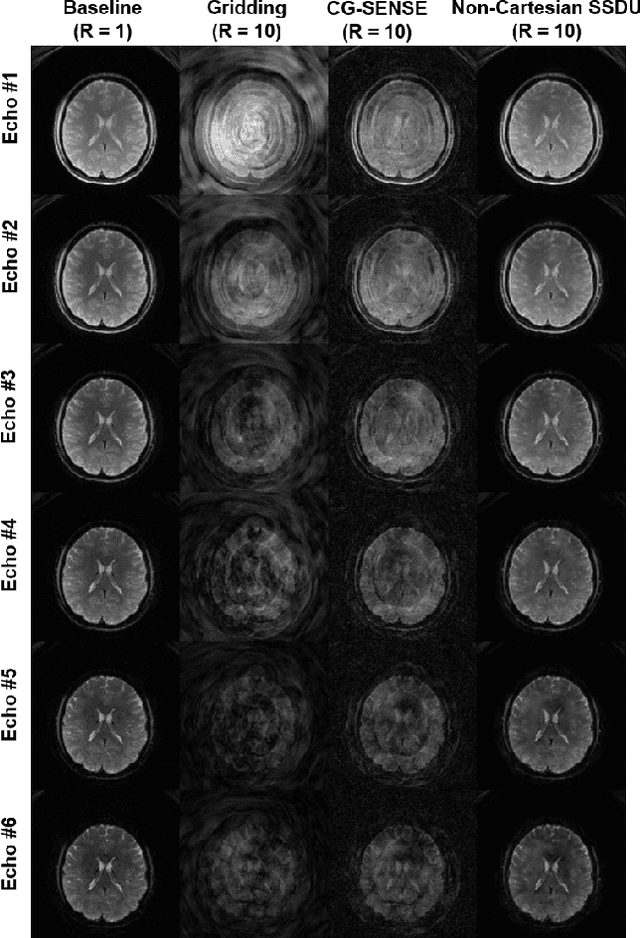
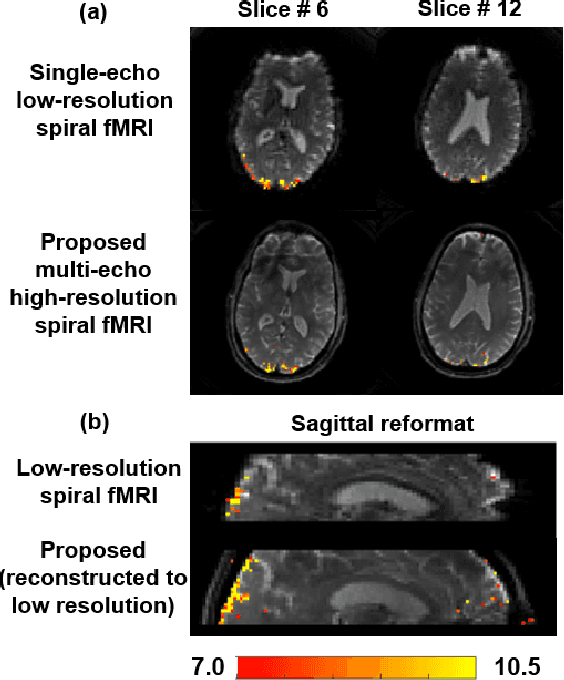
Abstract:Functional MRI (fMRI) is an important tool for non-invasive studies of brain function. Over the past decade, multi-echo fMRI methods that sample multiple echo times has become popular with potential to improve quantification. While these acquisitions are typically performed with Cartesian trajectories, non-Cartesian trajectories, in particular spiral acquisitions, hold promise for denser sampling of echo times. However, such acquisitions require very high acceleration rates for sufficient spatiotemporal resolutions. In this work, we propose to use a physics-driven deep learning (PD-DL) reconstruction to accelerate multi-echo spiral fMRI by 10-fold. We modify a self-supervised learning algorithm for optimized training with non-Cartesian trajectories and use it to train the PD-DL network. Results show that the proposed self-supervised PD-DL reconstruction achieves high spatio-temporal resolution with meaningful BOLD analysis.
On the Robustness of deep learning-based MRI Reconstruction to image transformations
Nov 21, 2022



Abstract:Although deep learning (DL) has received much attention in accelerated magnetic resonance imaging (MRI), recent studies show that tiny input perturbations may lead to instabilities of DL-based MRI reconstruction models. However, the approaches of robustifying these models are underdeveloped. Compared to image classification, it could be much more challenging to achieve a robust MRI image reconstruction network considering its regression-based learning objective, limited amount of training data, and lack of efficient robustness metrics. To circumvent the above limitations, our work revisits the problem of DL-based image reconstruction through the lens of robust machine learning. We find a new instability source of MRI image reconstruction, i.e., the lack of reconstruction robustness against spatial transformations of an input, e.g., rotation and cutout. Inspired by this new robustness metric, we develop a robustness-aware image reconstruction method that can defend against both pixel-wise adversarial perturbations as well as spatial transformations. Extensive experiments are also conducted to demonstrate the effectiveness of our proposed approaches.
Accelerated MRI With Deep Linear Convolutional Transform Learning
Apr 17, 2022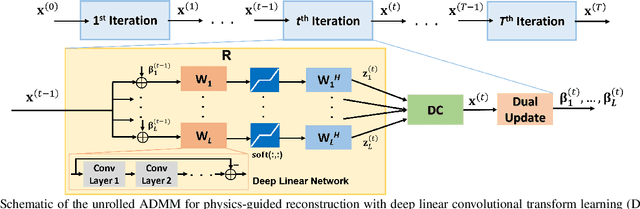
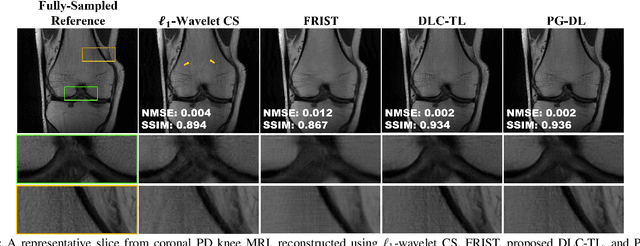
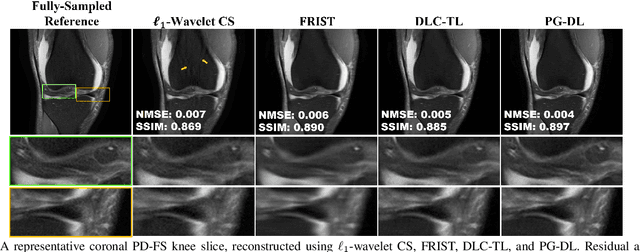
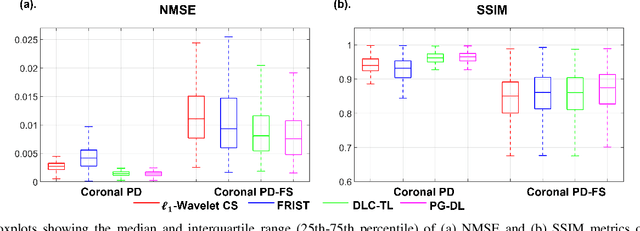
Abstract:Recent studies show that deep learning (DL) based MRI reconstruction outperforms conventional methods, such as parallel imaging and compressed sensing (CS), in multiple applications. Unlike CS that is typically implemented with pre-determined linear representations for regularization, DL inherently uses a non-linear representation learned from a large database. Another line of work uses transform learning (TL) to bridge the gap between these two approaches by learning linear representations from data. In this work, we combine ideas from CS, TL and DL reconstructions to learn deep linear convolutional transforms as part of an algorithm unrolling approach. Using end-to-end training, our results show that the proposed technique can reconstruct MR images to a level comparable to DL methods, while supporting uniform undersampling patterns unlike conventional CS methods. Our proposed method relies on convex sparse image reconstruction with linear representation at inference time, which may be beneficial for characterizing robustness, stability and generalizability.
Physics-Driven Deep Learning for Computational Magnetic Resonance Imaging
Mar 23, 2022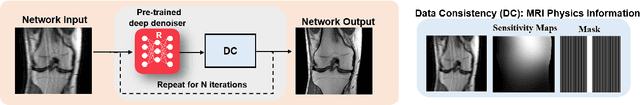
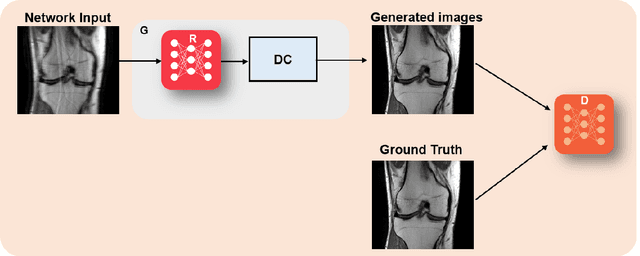
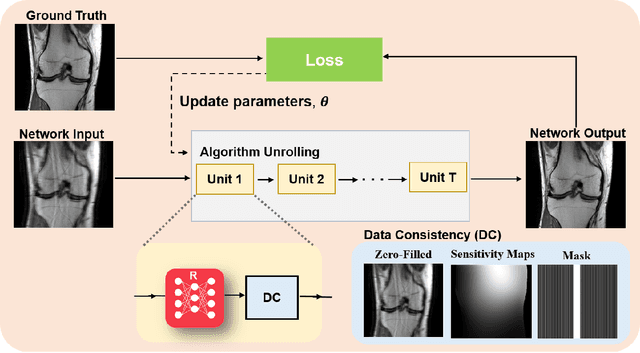
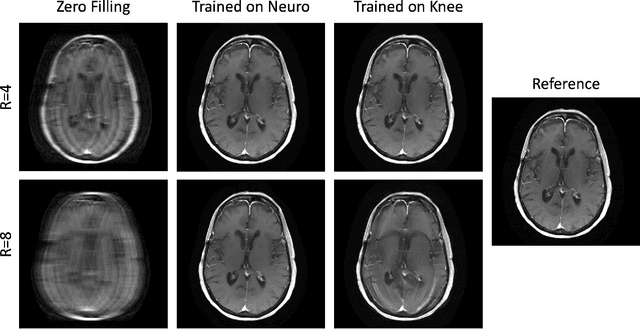
Abstract:Physics-driven deep learning methods have emerged as a powerful tool for computational magnetic resonance imaging (MRI) problems, pushing reconstruction performance to new limits. This article provides an overview of the recent developments in incorporating physics information into learning-based MRI reconstruction. We consider inverse problems with both linear and non-linear forward models for computational MRI, and review the classical approaches for solving these. We then focus on physics-driven deep learning approaches, covering physics-driven loss functions, plug-and-play methods, generative models, and unrolled networks. We highlight domain-specific challenges such as real- and complex-valued building blocks of neural networks, and translational applications in MRI with linear and non-linear forward models. Finally, we discuss common issues and open challenges, and draw connections to the importance of physics-driven learning when combined with other downstream tasks in the medical imaging pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge