Andreas Rauschecker
Mechanistic Learning with Guided Diffusion Models to Predict Spatio-Temporal Brain Tumor Growth
Sep 11, 2025Abstract:Predicting the spatio-temporal progression of brain tumors is essential for guiding clinical decisions in neuro-oncology. We propose a hybrid mechanistic learning framework that combines a mathematical tumor growth model with a guided denoising diffusion implicit model (DDIM) to synthesize anatomically feasible future MRIs from preceding scans. The mechanistic model, formulated as a system of ordinary differential equations, captures temporal tumor dynamics including radiotherapy effects and estimates future tumor burden. These estimates condition a gradient-guided DDIM, enabling image synthesis that aligns with both predicted growth and patient anatomy. We train our model on the BraTS adult and pediatric glioma datasets and evaluate on 60 axial slices of in-house longitudinal pediatric diffuse midline glioma (DMG) cases. Our framework generates realistic follow-up scans based on spatial similarity metrics. It also introduces tumor growth probability maps, which capture both clinically relevant extent and directionality of tumor growth as shown by 95th percentile Hausdorff Distance. The method enables biologically informed image generation in data-limited scenarios, offering generative-space-time predictions that account for mechanistic priors.
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
Domain Influence in MRI Medical Image Segmentation: spatial versus k-space inputs
Jul 01, 2024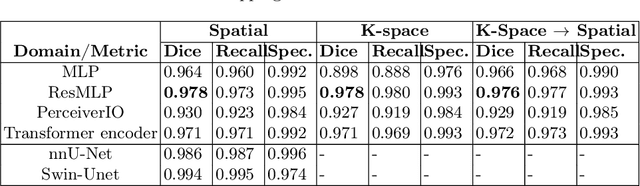


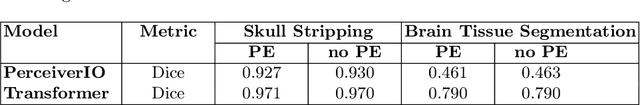
Abstract:Transformer-based networks applied to image patches have achieved cutting-edge performance in many vision tasks. However, lacking the built-in bias of convolutional neural networks (CNN) for local image statistics, they require large datasets and modifications to capture relationships between patches, especially in segmentation tasks. Images in the frequency domain might be more suitable for the attention mechanism, as local features are represented globally. By transforming images into the frequency domain, local features are represented globally. Due to MRI data acquisition properties, these images are particularly suitable. This work investigates how the image domain (spatial or k-space) affects segmentation results of deep learning (DL) models, focusing on attention-based networks and other non-convolutional models based on MLPs. We also examine the necessity of additional positional encoding for Transformer-based networks when input images are in the frequency domain. For evaluation, we pose a skull stripping task and a brain tissue segmentation task. The attention-based models used are PerceiverIO and a vanilla Transformer encoder. To compare with non-attention-based models, an MLP and ResMLP are also trained and tested. Results are compared with the Swin-Unet, the state-of-the-art medical image segmentation model. Experimental results show that using k-space for the input domain can significantly improve segmentation results. Also, additional positional encoding does not seem beneficial for attention-based networks if the input is in the frequency domain. Although none of the models matched the Swin-Unet's performance, the less complex models showed promising improvements with a different domain choice.
The University of California San Francisco Preoperative Diffuse Glioma MRI Dataset
Aug 30, 2021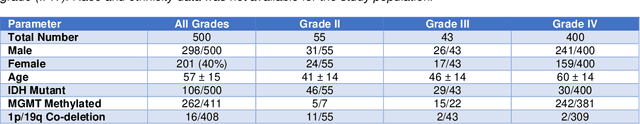

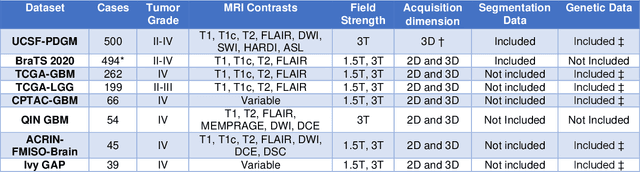
Abstract:Here we present the University of California San Francisco Preoperative Diffuse Glioma MRI (UCSF-PDGM) dataset. The UCSF-PDGM dataset includes 500 subjects with histopathologically-proven diffuse gliomas who were imaged with a standardized 3 Tesla preoperative brain tumor MRI protocol featuring predominantly 3D imaging, as well as advanced diffusion and perfusion imaging techniques. The dataset also includes isocitrate dehydrogenase (IDH) mutation status for all cases and O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation status for World Health Organization (WHO) grade III and IV gliomas. The UCSF-PDGM has been made publicly available in the hopes that researchers around the world will use these data to continue to push the boundaries of AI applications for diffuse gliomas.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge