Muhammad Ahmad Sultan
An unsupervised method for MRI recovery: Deep image prior with structured sparsity
Jan 02, 2025



Abstract:Objective: To propose and validate an unsupervised MRI reconstruction method that does not require fully sampled k-space data. Materials and Methods: The proposed method, deep image prior with structured sparsity (DISCUS), extends the deep image prior (DIP) by introducing group sparsity to frame-specific code vectors, enabling the discovery of a low-dimensional manifold for capturing temporal variations. \discus was validated using four studies: (I) simulation of a dynamic Shepp-Logan phantom to demonstrate its manifold discovery capabilities, (II) comparison with compressed sensing and DIP-based methods using simulated single-shot late gadolinium enhancement (LGE) image series from six distinct digital cardiac phantoms in terms of normalized mean square error (NMSE) and structural similarity index measure (SSIM), (III) evaluation on retrospectively undersampled single-shot LGE data from eight patients, and (IV) evaluation on prospectively undersampled single-shot LGE data from eight patients, assessed via blind scoring from two expert readers. Results: DISCUS outperformed competing methods, demonstrating superior reconstruction quality in terms of NMSE and SSIM (Studies I--III) and expert reader scoring (Study IV). Discussion: An unsupervised image reconstruction method is presented and validated on simulated and measured data. These developments can benefit applications where acquiring fully sampled data is challenging.
Motion-Guided Deep Image Prior for Cardiac MRI
Dec 05, 2024



Abstract:Cardiovascular magnetic resonance imaging is a powerful diagnostic tool for assessing cardiac structure and function. Traditional breath-held imaging protocols, however, pose challenges for patients with arrhythmias or limited breath-holding capacity. We introduce Motion-Guided Deep Image prior (M-DIP), a novel unsupervised reconstruction framework for accelerated real-time cardiac MRI. M-DIP employs a spatial dictionary to synthesize a time-dependent template image, which is further refined using time-dependent deformation fields that model cardiac and respiratory motion. Unlike prior DIP-based methods, M-DIP simultaneously captures physiological motion and frame-to-frame content variations, making it applicable to a wide range of dynamic applications. We validate M-DIP using simulated MRXCAT cine phantom data as well as free-breathing real-time cine and single-shot late gadolinium enhancement data from clinical patients. Comparative analyses against state-of-the-art supervised and unsupervised approaches demonstrate M-DIP's performance and versatility. M-DIP achieved better image quality metrics on phantom data, as well as higher reader scores for in-vivo patient data.
Deep Image prior with StruCtUred Sparsity for dynamic MRI reconstruction
Dec 01, 2023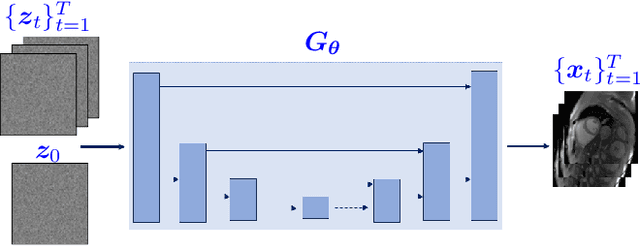
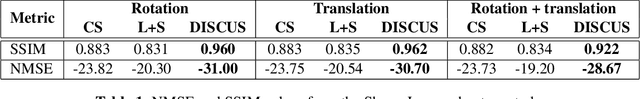
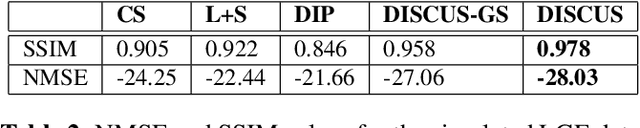
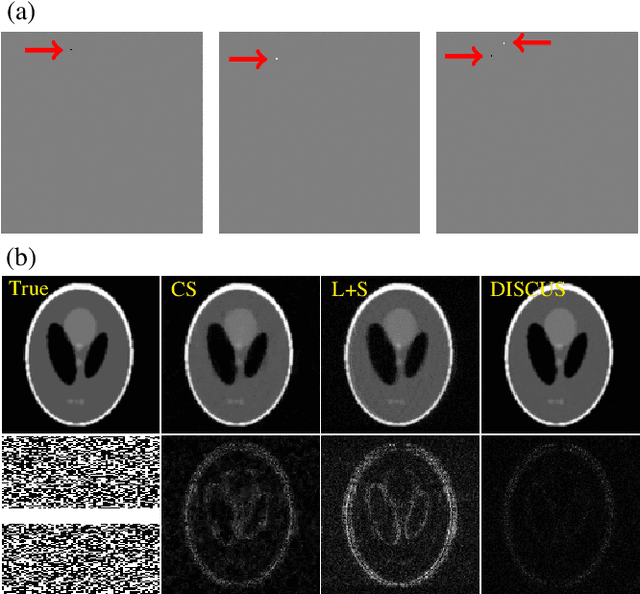
Abstract:High-quality training data are not always available in dynamic MRI. To address this, we propose a self-supervised deep learning method called deep image prior with structured sparsity (DISCUS) for reconstructing dynamic images. DISCUS is inspired by deep image prior (DIP) and recovers a series of images through joint optimization of network parameters and input code vectors. However, DISCUS additionally encourages group sparsity on frame-specific code vectors to discover the low-dimensional manifold that describes temporal variations across frames. Compared to prior work on manifold learning, DISCUS does not require specifying the manifold dimensionality. We validate DISCUS using three numerical studies. In the first study, we simulate a dynamic Shepp-Logan phantom with frames undergoing random rotations, translations, or both, and demonstrate that DISCUS can discover the dimensionality of the underlying manifold. In the second study, we use data from a realistic late gadolinium enhancement (LGE) phantom to compare DISCUS with compressed sensing (CS) and DIP and to demonstrate the positive impact of group sparsity. In the third study, we use retrospectively undersampled single-shot LGE data from five patients to compare DISCUS with CS reconstructions. The results from these studies demonstrate that DISCUS outperforms CS and DIP and that enforcing group sparsity on the code vectors helps discover true manifold dimensionality and provides additional performance gain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge