Laura Heacock
A Multi-Modal AI System for Screening Mammography: Integrating 2D and 3D Imaging to Improve Breast Cancer Detection in a Prospective Clinical Study
Apr 08, 2025
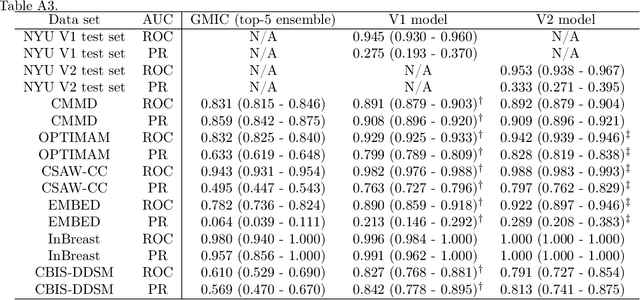
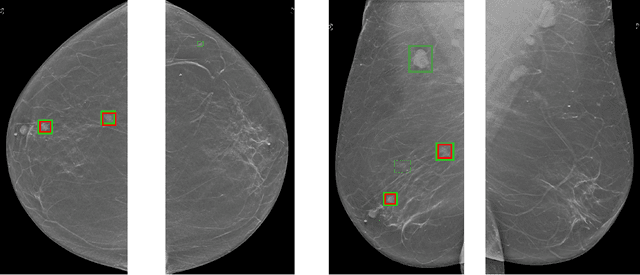
Abstract:Although digital breast tomosynthesis (DBT) improves diagnostic performance over full-field digital mammography (FFDM), false-positive recalls remain a concern in breast cancer screening. We developed a multi-modal artificial intelligence system integrating FFDM, synthetic mammography, and DBT to provide breast-level predictions and bounding-box localizations of suspicious findings. Our AI system, trained on approximately 500,000 mammography exams, achieved 0.945 AUROC on an internal test set. It demonstrated capacity to reduce recalls by 31.7% and radiologist workload by 43.8% while maintaining 100% sensitivity, underscoring its potential to improve clinical workflows. External validation confirmed strong generalizability, reducing the gap to a perfect AUROC by 35.31%-69.14% relative to strong baselines. In prospective deployment across 18 sites, the system reduced recall rates for low-risk cases. An improved version, trained on over 750,000 exams with additional labels, further reduced the gap by 18.86%-56.62% across large external datasets. Overall, these results underscore the importance of utilizing all available imaging modalities, demonstrate the potential for clinical impact, and indicate feasibility of further reduction of the test error with increased training set when using large-capacity neural networks.
A training regime to learn unified representations from complementary breast imaging modalities
Aug 16, 2024



Abstract:Full Field Digital Mammograms (FFDMs) and Digital Breast Tomosynthesis (DBT) are the two most widely used imaging modalities for breast cancer screening. Although DBT has increased cancer detection compared to FFDM, its widespread adoption in clinical practice has been slowed by increased interpretation times and a perceived decrease in the conspicuity of specific lesion types. Specifically, the non-inferiority of DBT for microcalcifications remains under debate. Due to concerns about the decrease in visual acuity, combined DBT-FFDM acquisitions remain popular, leading to overall increased exam times and radiation dosage. Enabling DBT to provide diagnostic information present in both FFDM and DBT would reduce reliance on FFDM, resulting in a reduction in both quantities. We propose a machine learning methodology that learns high-level representations leveraging the complementary diagnostic signal from both DBT and FFDM. Experiments on a large-scale data set validate our claims and show that our representations enable more accurate breast lesion detection than any DBT- or FFDM-based model.
fastMRI Breast: A publicly available radial k-space dataset of breast dynamic contrast-enhanced MRI
Jun 07, 2024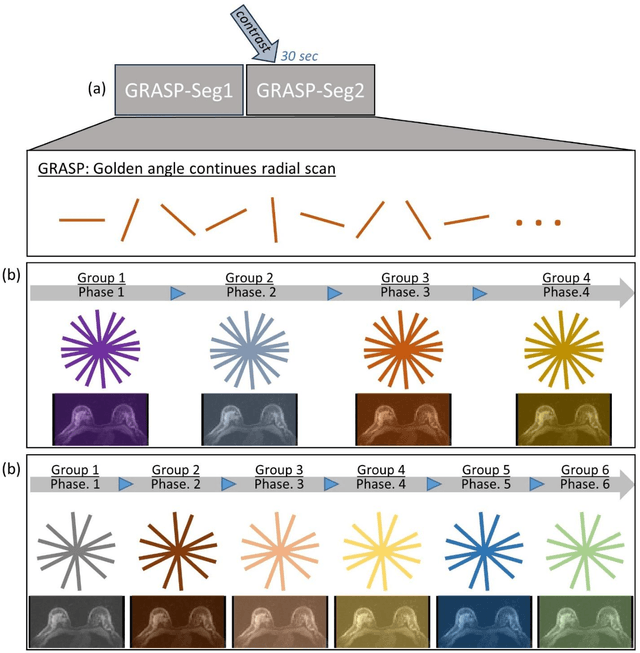
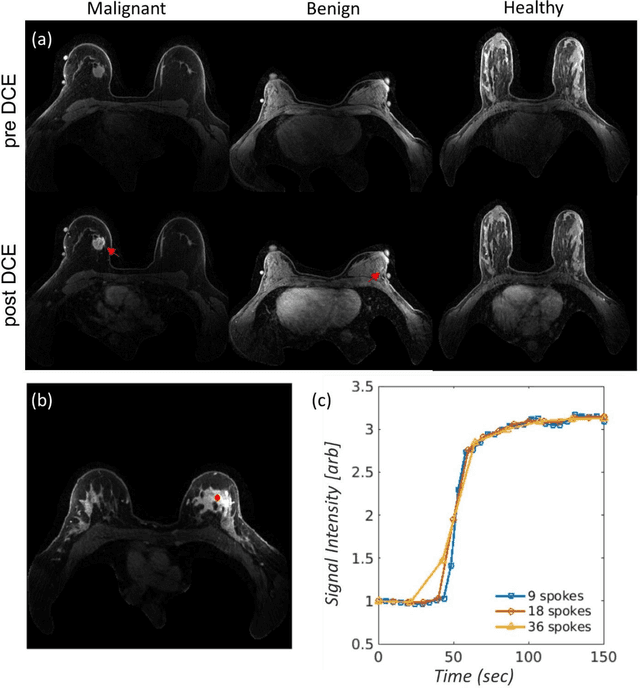
Abstract:This data curation work introduces the first large-scale dataset of radial k-space and DICOM data for breast DCE-MRI acquired in diagnostic breast MRI exams. Our dataset includes case-level labels indicating patient age, menopause status, lesion status (negative, benign, and malignant), and lesion type for each case. The public availability of this dataset and accompanying reconstruction code will support research and development of fast and quantitative breast image reconstruction and machine learning methods.
Understanding differences in applying DETR to natural and medical images
May 27, 2024Abstract:Transformer-based detectors have shown success in computer vision tasks with natural images. These models, exemplified by the Deformable DETR, are optimized through complex engineering strategies tailored to the typical characteristics of natural scenes. However, medical imaging data presents unique challenges such as extremely large image sizes, fewer and smaller regions of interest, and object classes which can be differentiated only through subtle differences. This study evaluates the applicability of these transformer-based design choices when applied to a screening mammography dataset that represents these distinct medical imaging data characteristics. Our analysis reveals that common design choices from the natural image domain, such as complex encoder architectures, multi-scale feature fusion, query initialization, and iterative bounding box refinement, do not improve and sometimes even impair object detection performance in medical imaging. In contrast, simpler and shallower architectures often achieve equal or superior results. This finding suggests that the adaptation of transformer models for medical imaging data requires a reevaluation of standard practices, potentially leading to more efficient and specialized frameworks for medical diagnosis.
Leveraging Transformers to Improve Breast Cancer Classification and Risk Assessment with Multi-modal and Longitudinal Data
Nov 15, 2023
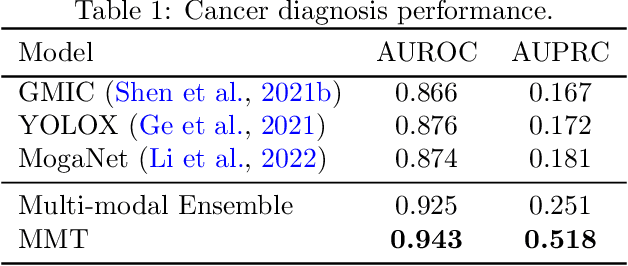
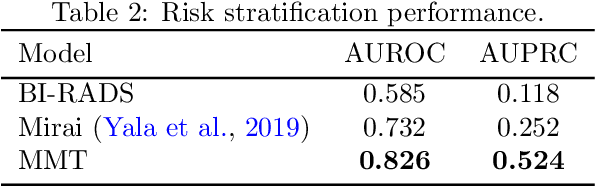
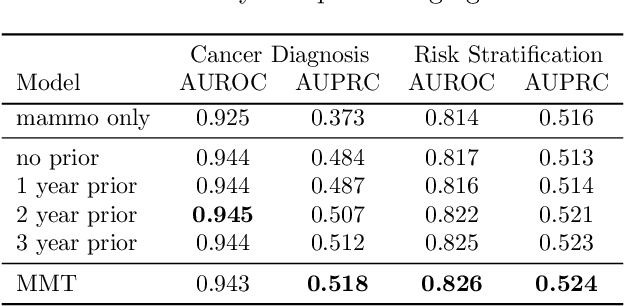
Abstract:Breast cancer screening, primarily conducted through mammography, is often supplemented with ultrasound for women with dense breast tissue. However, existing deep learning models analyze each modality independently, missing opportunities to integrate information across imaging modalities and time. In this study, we present Multi-modal Transformer (MMT), a neural network that utilizes mammography and ultrasound synergistically, to identify patients who currently have cancer and estimate the risk of future cancer for patients who are currently cancer-free. MMT aggregates multi-modal data through self-attention and tracks temporal tissue changes by comparing current exams to prior imaging. Trained on 1.3 million exams, MMT achieves an AUROC of 0.943 in detecting existing cancers, surpassing strong uni-modal baselines. For 5-year risk prediction, MMT attains an AUROC of 0.826, outperforming prior mammography-based risk models. Our research highlights the value of multi-modal and longitudinal imaging in cancer diagnosis and risk stratification.
BenchMD: A Benchmark for Modality-Agnostic Learning on Medical Images and Sensors
Apr 17, 2023



Abstract:Medical data poses a daunting challenge for AI algorithms: it exists in many different modalities, experiences frequent distribution shifts, and suffers from a scarcity of examples and labels. Recent advances, including transformers and self-supervised learning, promise a more universal approach that can be applied flexibly across these diverse conditions. To measure and drive progress in this direction, we present BenchMD: a benchmark that tests how modality-agnostic methods, including architectures and training techniques (e.g. self-supervised learning, ImageNet pretraining), perform on a diverse array of clinically-relevant medical tasks. BenchMD combines 19 publicly available datasets for 7 medical modalities, including 1D sensor data, 2D images, and 3D volumetric scans. Our benchmark reflects real-world data constraints by evaluating methods across a range of dataset sizes, including challenging few-shot settings that incentivize the use of pretraining. Finally, we evaluate performance on out-of-distribution data collected at different hospitals than the training data, representing naturally-occurring distribution shifts that frequently degrade the performance of medical AI models. Our baseline results demonstrate that no modality-agnostic technique achieves strong performance across all modalities, leaving ample room for improvement on the benchmark. Code is released at https://github.com/rajpurkarlab/BenchMD .
3D-GMIC: an efficient deep neural network to find small objects in large 3D images
Oct 16, 2022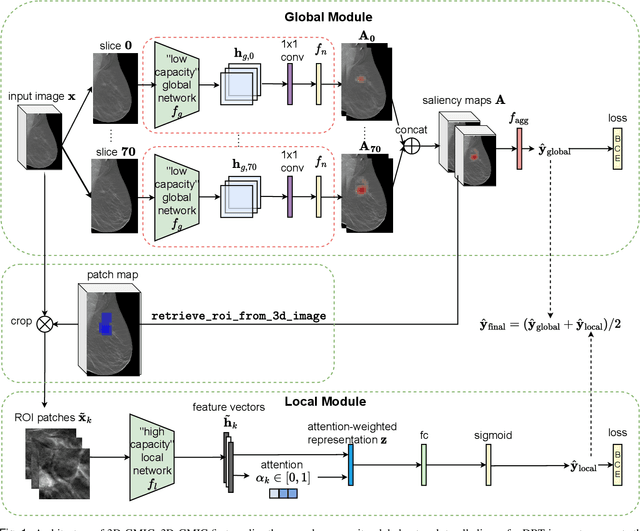
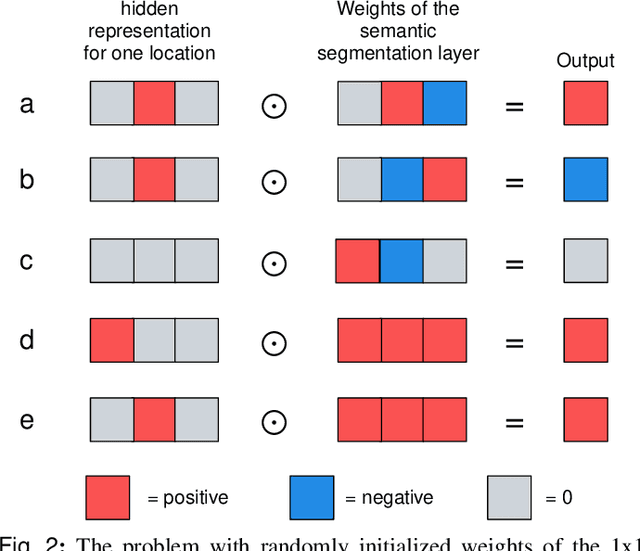
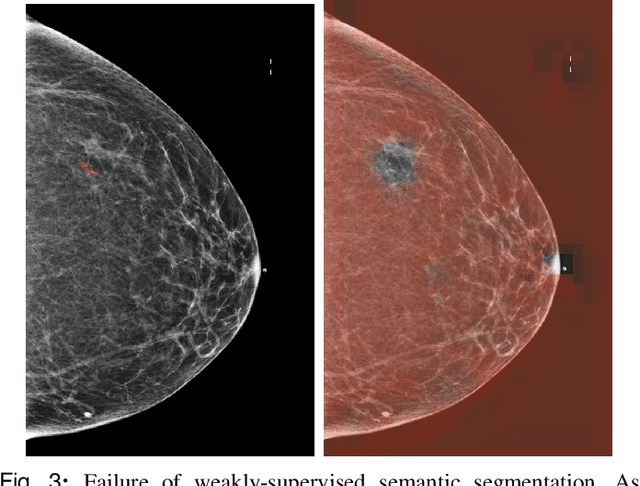
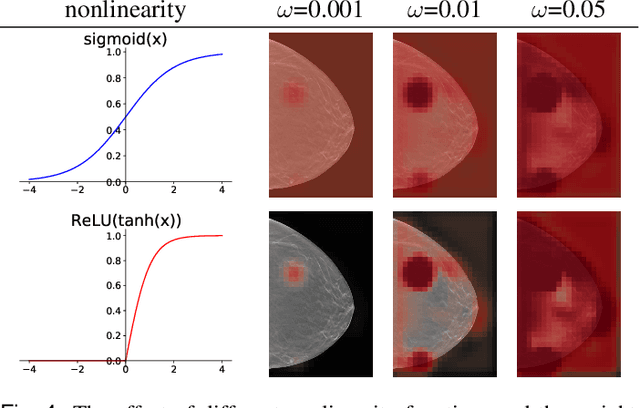
Abstract:3D imaging enables a more accurate diagnosis by providing spatial information about organ anatomy. However, using 3D images to train AI models is computationally challenging because they consist of tens or hundreds of times more pixels than their 2D counterparts. To train with high-resolution 3D images, convolutional neural networks typically resort to downsampling them or projecting them to two dimensions. In this work, we propose an effective alternative, a novel neural network architecture that enables computationally efficient classification of 3D medical images in their full resolution. Compared to off-the-shelf convolutional neural networks, 3D-GMIC uses 77.98%-90.05% less GPU memory and 91.23%-96.02% less computation. While our network is trained only with image-level labels, without segmentation labels, it explains its classification predictions by providing pixel-level saliency maps. On a dataset collected at NYU Langone Health, including 85,526 patients with full-field 2D mammography (FFDM), synthetic 2D mammography, and 3D mammography (DBT), our model, the 3D Globally-Aware Multiple Instance Classifier (3D-GMIC), achieves a breast-wise AUC of 0.831 (95% CI: 0.769-0.887) in classifying breasts with malignant findings using DBT images. As DBT and 2D mammography capture different information, averaging predictions on 2D and 3D mammography together leads to a diverse ensemble with an improved breast-wise AUC of 0.841 (95% CI: 0.768-0.895). Our model generalizes well to an external dataset from Duke University Hospital, achieving an image-wise AUC of 0.848 (95% CI: 0.798-0.896) in classifying DBT images with malignant findings.
Differences between human and machine perception in medical diagnosis
Nov 28, 2020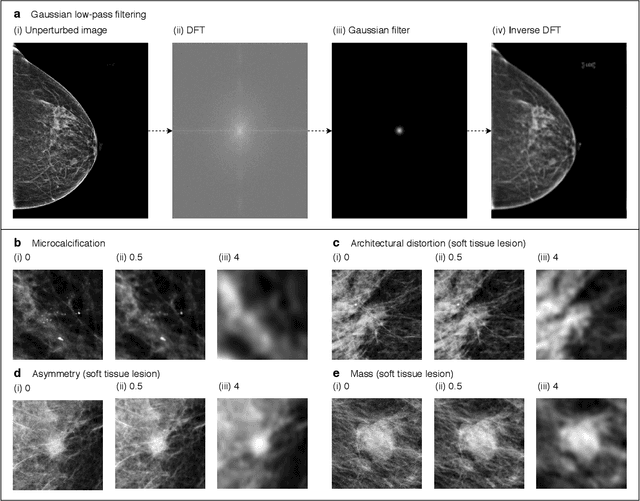
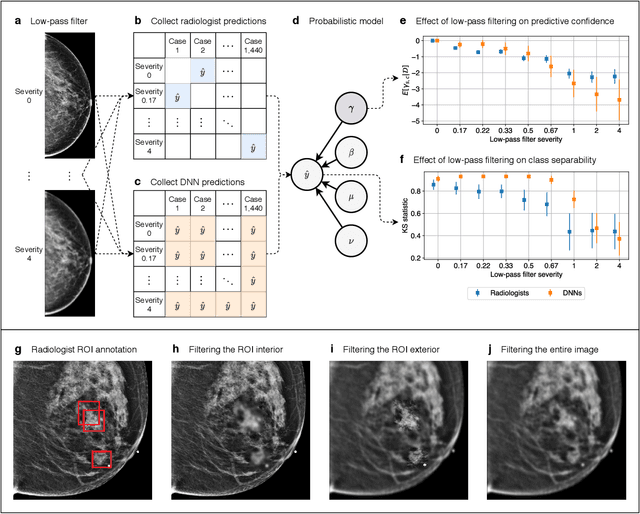
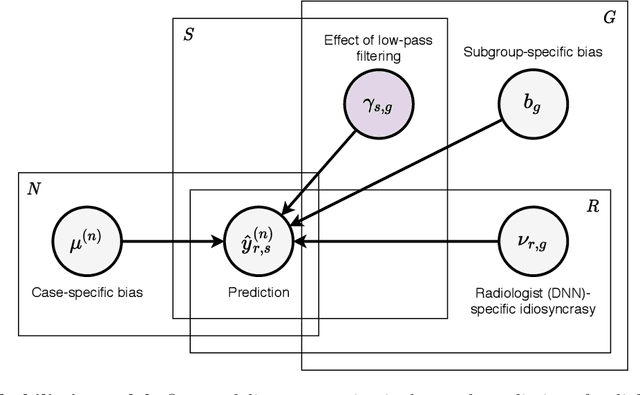
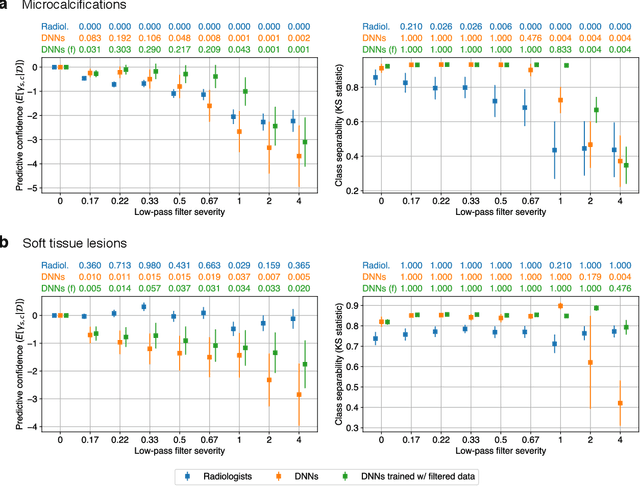
Abstract:Deep neural networks (DNNs) show promise in image-based medical diagnosis, but cannot be fully trusted since their performance can be severely degraded by dataset shifts to which human perception remains invariant. If we can better understand the differences between human and machine perception, we can potentially characterize and mitigate this effect. We therefore propose a framework for comparing human and machine perception in medical diagnosis. The two are compared with respect to their sensitivity to the removal of clinically meaningful information, and to the regions of an image deemed most suspicious. Drawing inspiration from the natural image domain, we frame both comparisons in terms of perturbation robustness. The novelty of our framework is that separate analyses are performed for subgroups with clinically meaningful differences. We argue that this is necessary in order to avert Simpson's paradox and draw correct conclusions. We demonstrate our framework with a case study in breast cancer screening, and reveal significant differences between radiologists and DNNs. We compare the two with respect to their robustness to Gaussian low-pass filtering, performing a subgroup analysis on microcalcifications and soft tissue lesions. For microcalcifications, DNNs use a separate set of high frequency components than radiologists, some of which lie outside the image regions considered most suspicious by radiologists. These features run the risk of being spurious, but if not, could represent potential new biomarkers. For soft tissue lesions, the divergence between radiologists and DNNs is even starker, with DNNs relying heavily on spurious high frequency components ignored by radiologists. Importantly, this deviation in soft tissue lesions was only observable through subgroup analysis, which highlights the importance of incorporating medical domain knowledge into our comparison framework.
Reducing false-positive biopsies with deep neural networks that utilize local and global information in screening mammograms
Sep 19, 2020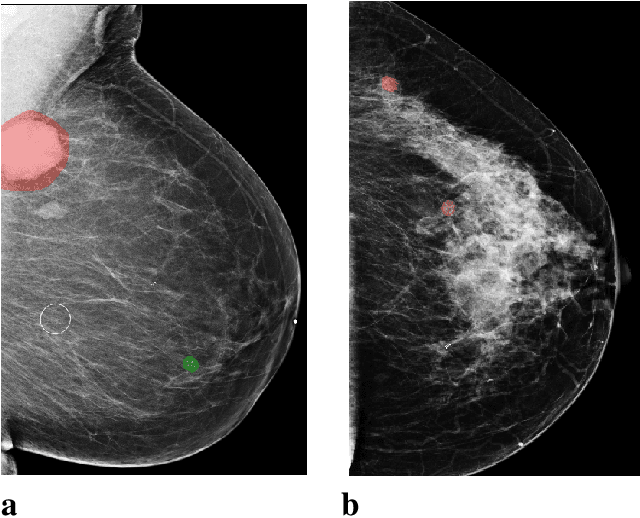
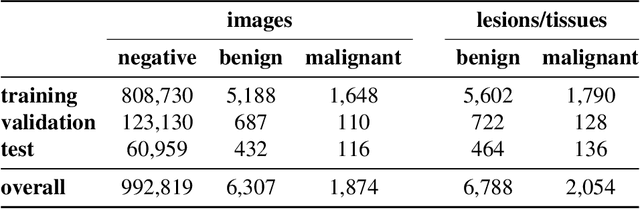
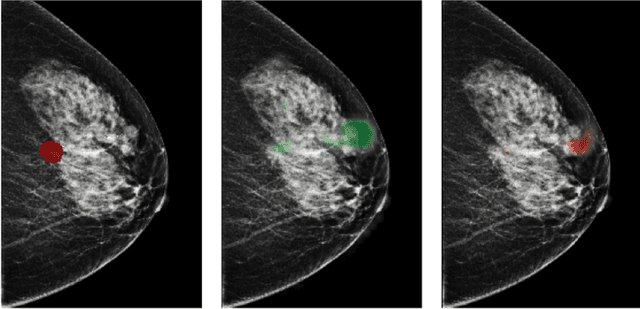
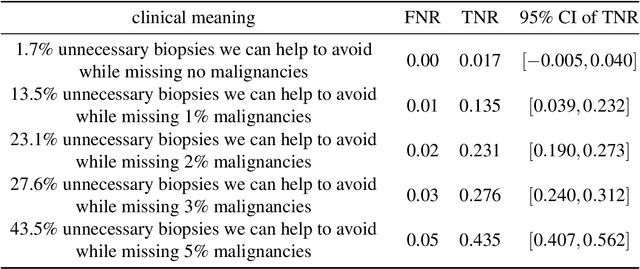
Abstract:Breast cancer is the most common cancer in women, and hundreds of thousands of unnecessary biopsies are done around the world at a tremendous cost. It is crucial to reduce the rate of biopsies that turn out to be benign tissue. In this study, we build deep neural networks (DNNs) to classify biopsied lesions as being either malignant or benign, with the goal of using these networks as second readers serving radiologists to further reduce the number of false positive findings. We enhance the performance of DNNs that are trained to learn from small image patches by integrating global context provided in the form of saliency maps learned from the entire image into their reasoning, similar to how radiologists consider global context when evaluating areas of interest. Our experiments are conducted on a dataset of 229,426 screening mammography exams from 141,473 patients. We achieve an AUC of 0.8 on a test set consisting of 464 benign and 136 malignant lesions.
Understanding the robustness of deep neural network classifiers for breast cancer screening
Mar 23, 2020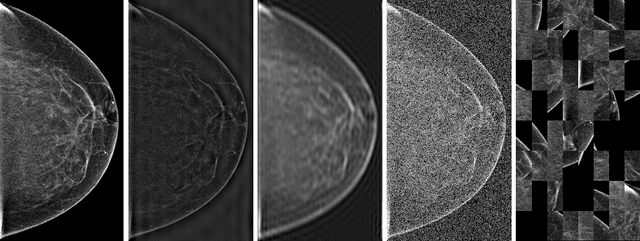
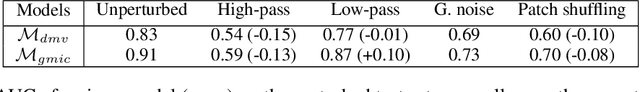
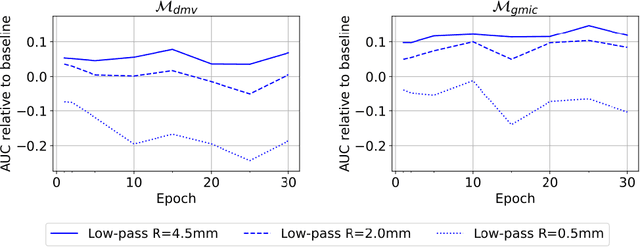
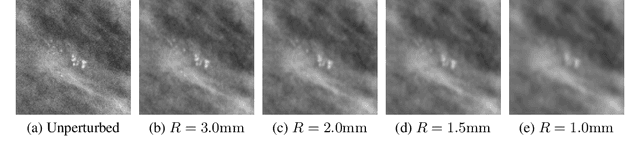
Abstract:Deep neural networks (DNNs) show promise in breast cancer screening, but their robustness to input perturbations must be better understood before they can be clinically implemented. There exists extensive literature on this subject in the context of natural images that can potentially be built upon. However, it cannot be assumed that conclusions about robustness will transfer from natural images to mammogram images, due to significant differences between the two image modalities. In order to determine whether conclusions will transfer, we measure the sensitivity of a radiologist-level screening mammogram image classifier to four commonly studied input perturbations that natural image classifiers are sensitive to. We find that mammogram image classifiers are also sensitive to these perturbations, which suggests that we can build on the existing literature. We also perform a detailed analysis on the effects of low-pass filtering, and find that it degrades the visibility of clinically meaningful features called microcalcifications. Since low-pass filtering removes semantically meaningful information that is predictive of breast cancer, we argue that it is undesirable for mammogram image classifiers to be invariant to it. This is in contrast to natural images, where we do not want DNNs to be sensitive to low-pass filtering due to its tendency to remove information that is human-incomprehensible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge