Carlos Fernandez-Granda
Atomic Depth Estimation From Noisy Electron Microscopy Data Via Deep Learning
Jan 27, 2026Abstract:We present a novel approach for extracting 3D atomic-level information from transmission electron microscopy (TEM) images affected by significant noise. The approach is based on formulating depth estimation as a semantic segmentation problem. We address the resulting segmentation problem by training a deep convolutional neural network to generate pixel-wise depth segmentation maps using simulated data corrupted by synthetic noise. The proposed method was applied to estimate the depth of atomic columns in CeO2 nanoparticles from simulated images and real-world TEM data. Our experiments show that the resulting depth estimates are accurate, calibrated and robust to noise.
SamudrACE: Fast and Accurate Coupled Climate Modeling with 3D Ocean and Atmosphere Emulators
Sep 15, 2025
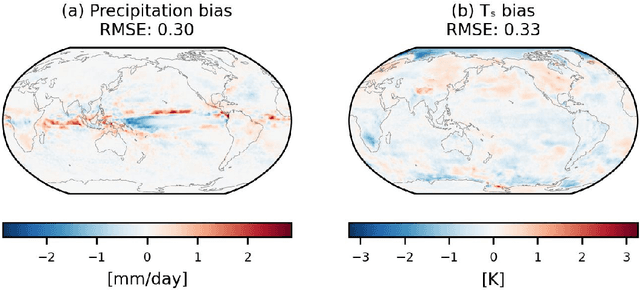


Abstract:Traditional numerical global climate models simulate the full Earth system by exchanging boundary conditions between separate simulators of the atmosphere, ocean, sea ice, land surface, and other geophysical processes. This paradigm allows for distributed development of individual components within a common framework, unified by a coupler that handles translation between realms via spatial or temporal alignment and flux exchange. Following a similar approach adapted for machine learning-based emulators, we present SamudrACE: a coupled global climate model emulator which produces centuries-long simulations at 1-degree horizontal, 6-hourly atmospheric, and 5-daily oceanic resolution, with 145 2D fields spanning 8 atmospheric and 19 oceanic vertical levels, plus sea ice, surface, and top-of-atmosphere variables. SamudrACE is highly stable and has low climate biases comparable to those of its components with prescribed boundary forcing, with realistic variability in coupled climate phenomena such as ENSO that is not possible to simulate in uncoupled mode.
Black Box Causal Inference: Effect Estimation via Meta Prediction
Mar 07, 2025



Abstract:Causal inference and the estimation of causal effects plays a central role in decision-making across many areas, including healthcare and economics. Estimating causal effects typically requires an estimator that is tailored to each problem of interest. But developing estimators can take significant effort for even a single causal inference setting. For example, algorithms for regression-based estimators, propensity score methods, and doubly robust methods were designed across several decades to handle causal estimation with observed confounders. Similarly, several estimators have been developed to exploit instrumental variables (IVs), including two-stage least-squares (TSLS), control functions, and the method-of-moments. In this work, we instead frame causal inference as a dataset-level prediction problem, offloading algorithm design to the learning process. The approach we introduce, called black box causal inference (BBCI), builds estimators in a black-box manner by learning to predict causal effects from sampled dataset-effect pairs. We demonstrate accurate estimation of average treatment effects (ATEs) and conditional average treatment effects (CATEs) with BBCI across several causal inference problems with known identification, including problems with less developed estimators.
Samudra: An AI Global Ocean Emulator for Climate
Dec 05, 2024

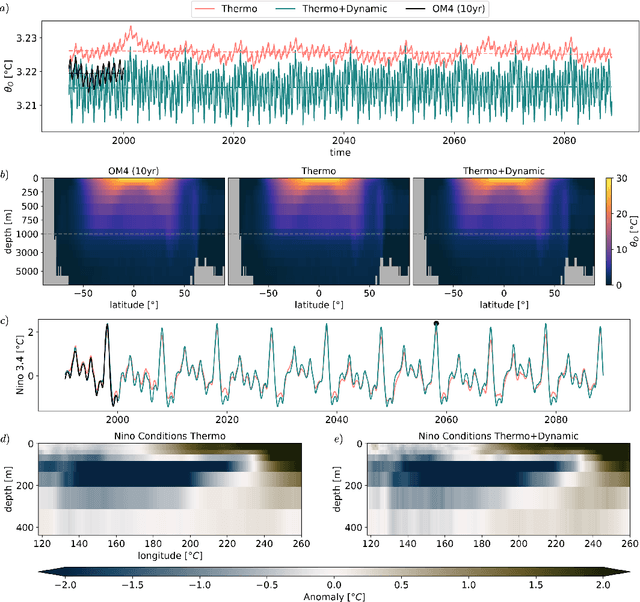
Abstract:AI emulators for forecasting have emerged as powerful tools that can outperform conventional numerical predictions. The next frontier is to build emulators for long-term climate projections with robust skill across a wide range of spatiotemporal scales, a particularly important goal for the ocean. Our work builds a skillful global emulator of the ocean component of a state-of-the-art climate model. We emulate key ocean variables, sea surface height, horizontal velocities, temperature, and salinity, across their full depth. We use a modified ConvNeXt UNet architecture trained on multidepth levels of ocean data. We show that the ocean emulator - Samudra - which exhibits no drift relative to the truth, can reproduce the depth structure of ocean variables and their interannual variability. Samudra is stable for centuries and 150 times faster than the original ocean model. Samudra struggles to capture the correct magnitude of the forcing trends and simultaneously remains stable, requiring further work.
Active learning for efficient discovery of optimal gene combinations in the combinatorial perturbation space
Nov 18, 2024



Abstract:The advancement of novel combinatorial CRISPR screening technologies enables the identification of synergistic gene combinations on a large scale. This is crucial for developing novel and effective combination therapies, but the combinatorial space makes exhaustive experimentation infeasible. We introduce NAIAD, an active learning framework that efficiently discovers optimal gene pairs capable of driving cells toward desired cellular phenotypes. NAIAD leverages single-gene perturbation effects and adaptive gene embeddings that scale with the training data size, mitigating overfitting in small-sample learning while capturing complex gene interactions as more data is collected. Evaluated on four CRISPR combinatorial perturbation datasets totaling over 350,000 genetic interactions, NAIAD, trained on small datasets, outperforms existing models by up to 40\% relative to the second-best. NAIAD's recommendation system prioritizes gene pairs with the maximum predicted effects, resulting in the highest marginal gain in each AI-experiment round and accelerating discovery with fewer CRISPR experimental iterations. Our NAIAD framework (https://github.com/NeptuneBio/NAIAD) improves the identification of novel, effective gene combinations, enabling more efficient CRISPR library design and offering promising applications in genomics research and therapeutic development.
A Monte Carlo Framework for Calibrated Uncertainty Estimation in Sequence Prediction
Oct 30, 2024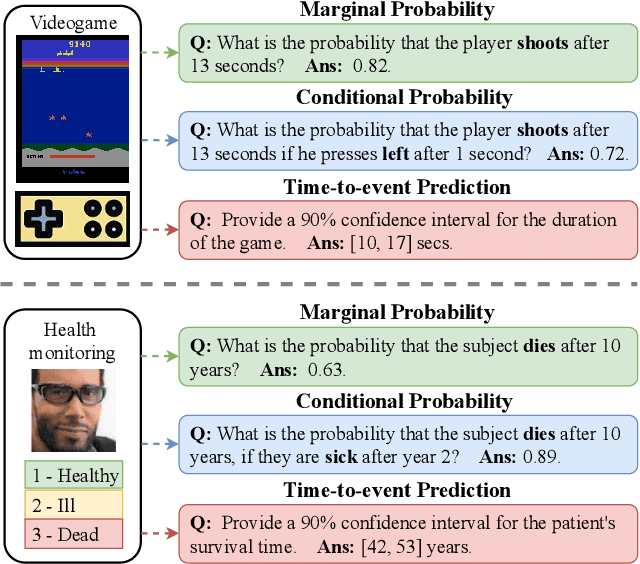

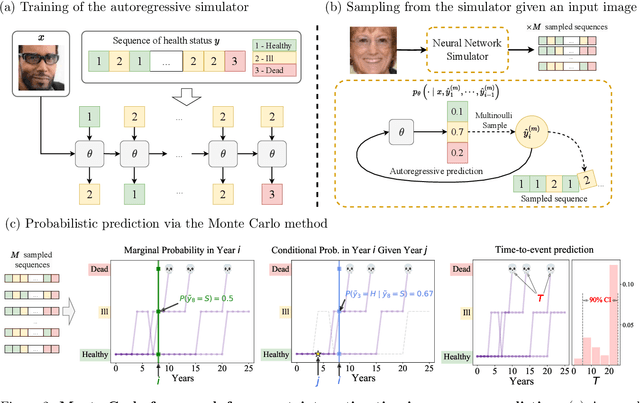

Abstract:Probabilistic prediction of sequences from images and other high-dimensional data is a key challenge, particularly in risk-sensitive applications. In these settings, it is often desirable to quantify the uncertainty associated with the prediction (instead of just determining the most likely sequence, as in language modeling). In this paper, we propose a Monte Carlo framework to estimate probabilities and confidence intervals associated with the distribution of a discrete sequence. Our framework uses a Monte Carlo simulator, implemented as an autoregressively trained neural network, to sample sequences conditioned on an image input. We then use these samples to estimate the probabilities and confidence intervals. Experiments on synthetic and real data show that the framework produces accurate discriminative predictions, but can suffer from miscalibration. In order to address this shortcoming, we propose a time-dependent regularization method, which is shown to produce calibrated predictions.
Multi-modal AI for comprehensive breast cancer prognostication
Oct 28, 2024Abstract:Treatment selection in breast cancer is guided by molecular subtypes and clinical characteristics. Recurrence risk assessment plays a crucial role in personalizing treatment. Current methods, including genomic assays, have limited accuracy and clinical utility, leading to suboptimal decisions for many patients. We developed a test for breast cancer patient stratification based on digital pathology and clinical characteristics using novel AI methods. Specifically, we utilized a vision transformer-based pan-cancer foundation model trained with self-supervised learning to extract features from digitized H&E-stained slides. These features were integrated with clinical data to form a multi-modal AI test predicting cancer recurrence and death. The test was developed and evaluated using data from a total of 8,161 breast cancer patients across 15 cohorts originating from seven countries. Of these, 3,502 patients from five cohorts were used exclusively for evaluation, while the remaining patients were used for training. Our test accurately predicted our primary endpoint, disease-free interval, in the five external cohorts (C-index: 0.71 [0.68-0.75], HR: 3.63 [3.02-4.37, p<0.01]). In a direct comparison (N=858), the AI test was more accurate than Oncotype DX, the standard-of-care 21-gene assay, with a C-index of 0.67 [0.61-0.74] versus 0.61 [0.49-0.73], respectively. Additionally, the AI test added independent information to Oncotype DX in a multivariate analysis (HR: 3.11 [1.91-5.09, p<0.01)]). The test demonstrated robust accuracy across all major breast cancer subtypes, including TNBC (C-index: 0.71 [0.62-0.81], HR: 3.81 [2.35-6.17, p=0.02]), where no diagnostic tools are currently recommended by clinical guidelines. These results suggest that our AI test can improve accuracy, extend applicability to a wider range of patients, and enhance access to treatment selection tools.
Understanding differences in applying DETR to natural and medical images
May 27, 2024Abstract:Transformer-based detectors have shown success in computer vision tasks with natural images. These models, exemplified by the Deformable DETR, are optimized through complex engineering strategies tailored to the typical characteristics of natural scenes. However, medical imaging data presents unique challenges such as extremely large image sizes, fewer and smaller regions of interest, and object classes which can be differentiated only through subtle differences. This study evaluates the applicability of these transformer-based design choices when applied to a screening mammography dataset that represents these distinct medical imaging data characteristics. Our analysis reveals that common design choices from the natural image domain, such as complex encoder architectures, multi-scale feature fusion, query initialization, and iterative bounding box refinement, do not improve and sometimes even impair object detection performance in medical imaging. In contrast, simpler and shallower architectures often achieve equal or superior results. This finding suggests that the adaptation of transformer models for medical imaging data requires a reevaluation of standard practices, potentially leading to more efficient and specialized frameworks for medical diagnosis.
Making Self-supervised Learning Robust to Spurious Correlation via Learning-speed Aware Sampling
Nov 29, 2023Abstract:Self-supervised learning (SSL) has emerged as a powerful technique for learning rich representations from unlabeled data. The data representations are able to capture many underlying attributes of data, and be useful in downstream prediction tasks. In real-world settings, spurious correlations between some attributes (e.g. race, gender and age) and labels for downstream tasks often exist, e.g. cancer is usually more prevalent among elderly patients. In this paper, we investigate SSL in the presence of spurious correlations and show that the SSL training loss can be minimized by capturing only a subset of the conspicuous features relevant to those sensitive attributes, despite the presence of other important predictive features for the downstream tasks. To address this issue, we investigate the learning dynamics of SSL and observe that the learning is slower for samples that conflict with such correlations (e.g. elder patients without cancer). Motivated by these findings, we propose a learning-speed aware SSL (LA-SSL) approach, in which we sample each training data with a probability that is inversely related to its learning speed. We evaluate LA-SSL on three datasets that exhibit spurious correlations between different attributes, demonstrating that it improves the robustness of pretrained representations on downstream classification tasks.
Quantifying Impairment and Disease Severity Using AI Models Trained on Healthy Subjects
Nov 21, 2023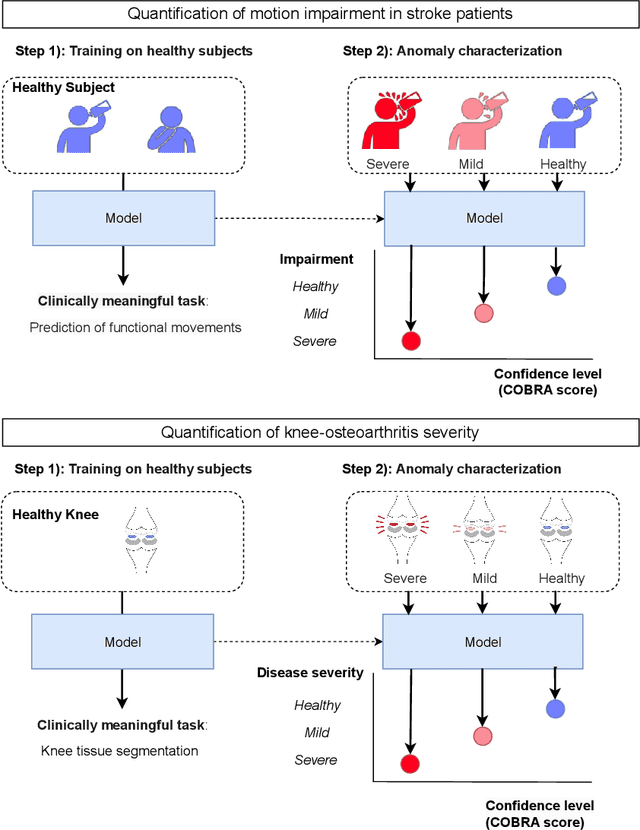
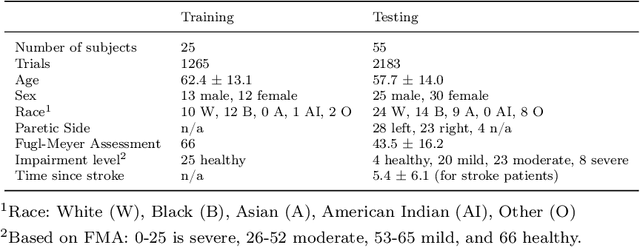
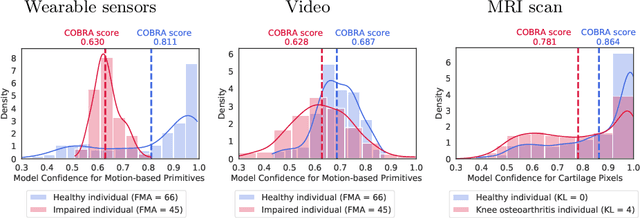
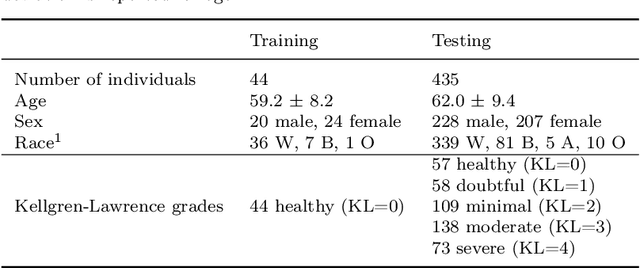
Abstract:Automatic assessment of impairment and disease severity is a key challenge in data-driven medicine. We propose a novel framework to address this challenge, which leverages AI models trained exclusively on healthy individuals. The COnfidence-Based chaRacterization of Anomalies (COBRA) score exploits the decrease in confidence of these models when presented with impaired or diseased patients to quantify their deviation from the healthy population. We applied the COBRA score to address a key limitation of current clinical evaluation of upper-body impairment in stroke patients. The gold-standard Fugl-Meyer Assessment (FMA) requires in-person administration by a trained assessor for 30-45 minutes, which restricts monitoring frequency and precludes physicians from adapting rehabilitation protocols to the progress of each patient. The COBRA score, computed automatically in under one minute, is shown to be strongly correlated with the FMA on an independent test cohort for two different data modalities: wearable sensors ($\rho = 0.845$, 95% CI [0.743,0.908]) and video ($\rho = 0.746$, 95% C.I [0.594, 0.847]). To demonstrate the generalizability of the approach to other conditions, the COBRA score was also applied to quantify severity of knee osteoarthritis from magnetic-resonance imaging scans, again achieving significant correlation with an independent clinical assessment ($\rho = 0.644$, 95% C.I [0.585,0.696]).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge