Kangning Liu
Alex
Sparse-LaViDa: Sparse Multimodal Discrete Diffusion Language Models
Dec 16, 2025Abstract:Masked Discrete Diffusion Models (MDMs) have achieved strong performance across a wide range of multimodal tasks, including image understanding, generation, and editing. However, their inference speed remains suboptimal due to the need to repeatedly process redundant masked tokens at every sampling step. In this work, we propose Sparse-LaViDa, a novel modeling framework that dynamically truncates unnecessary masked tokens at each inference step to accelerate MDM sampling. To preserve generation quality, we introduce specialized register tokens that serve as compact representations for the truncated tokens. Furthermore, to ensure consistency between training and inference, we design a specialized attention mask that faithfully matches the truncated sampling procedure during training. Built upon the state-of-the-art unified MDM LaViDa-O, Sparse-LaViDa achieves up to a 2x speedup across diverse tasks including text-to-image generation, image editing, and mathematical reasoning, while maintaining generation quality.
VGent: Visual Grounding via Modular Design for Disentangling Reasoning and Prediction
Dec 11, 2025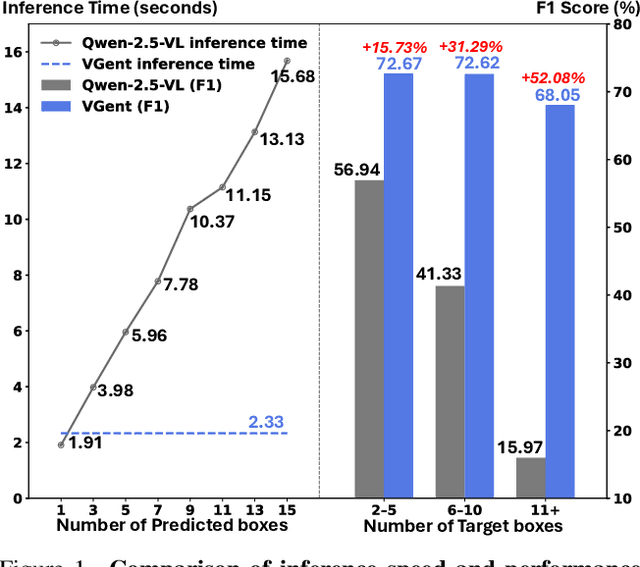
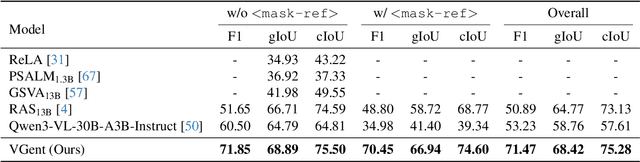
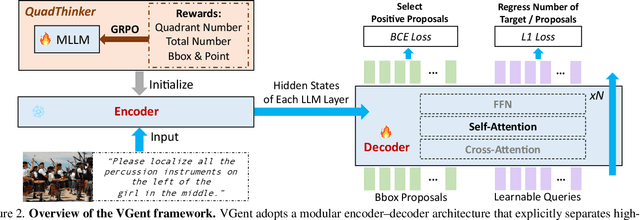
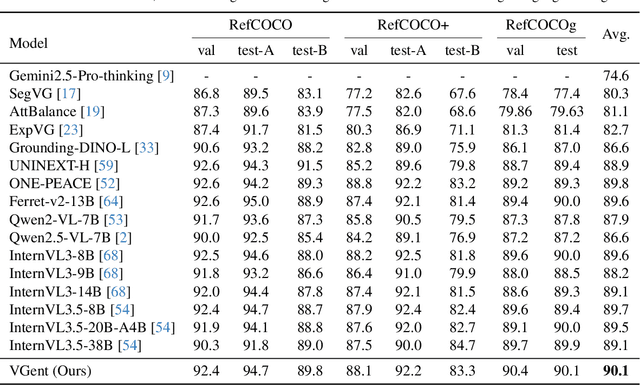
Abstract:Current visual grounding models are either based on a Multimodal Large Language Model (MLLM) that performs auto-regressive decoding, which is slow and risks hallucinations, or on re-aligning an LLM with vision features to learn new special or object tokens for grounding, which may undermine the LLM's pretrained reasoning ability. In contrast, we propose VGent, a modular encoder-decoder architecture that explicitly disentangles high-level reasoning and low-level bounding box prediction. Specifically, a frozen MLLM serves as the encoder to provide untouched powerful reasoning capabilities, while a decoder takes high-quality boxes proposed by detectors as queries and selects target box(es) via cross-attending on encoder's hidden states. This design fully leverages advances in both object detection and MLLM, avoids the pitfalls of auto-regressive decoding, and enables fast inference. Moreover, it supports modular upgrades of both the encoder and decoder to benefit the whole system: we introduce (i) QuadThinker, an RL-based training paradigm for enhancing multi-target reasoning ability of the encoder; (ii) mask-aware label for resolving detection-segmentation ambiguity; and (iii) global target recognition to improve the recognition of all the targets which benefits the selection among augmented proposals. Experiments on multi-target visual grounding benchmarks show that VGent achieves a new state-of-the-art with +20.6% F1 improvement over prior methods, and further boosts gIoU by +8.2% and cIoU by +5.8% under visual reference challenges, while maintaining constant, fast inference latency.
Refer to Anything with Vision-Language Prompts
Jun 05, 2025Abstract:Recent image segmentation models have advanced to segment images into high-quality masks for visual entities, and yet they cannot provide comprehensive semantic understanding for complex queries based on both language and vision. This limitation reduces their effectiveness in applications that require user-friendly interactions driven by vision-language prompts. To bridge this gap, we introduce a novel task of omnimodal referring expression segmentation (ORES). In this task, a model produces a group of masks based on arbitrary prompts specified by text only or text plus reference visual entities. To address this new challenge, we propose a novel framework to "Refer to Any Segmentation Mask Group" (RAS), which augments segmentation models with complex multimodal interactions and comprehension via a mask-centric large multimodal model. For training and benchmarking ORES models, we create datasets MaskGroups-2M and MaskGroups-HQ to include diverse mask groups specified by text and reference entities. Through extensive evaluation, we demonstrate superior performance of RAS on our new ORES task, as well as classic referring expression segmentation (RES) and generalized referring expression segmentation (GRES) tasks. Project page: https://Ref2Any.github.io.
Multi-modal AI for comprehensive breast cancer prognostication
Oct 28, 2024Abstract:Treatment selection in breast cancer is guided by molecular subtypes and clinical characteristics. Recurrence risk assessment plays a crucial role in personalizing treatment. Current methods, including genomic assays, have limited accuracy and clinical utility, leading to suboptimal decisions for many patients. We developed a test for breast cancer patient stratification based on digital pathology and clinical characteristics using novel AI methods. Specifically, we utilized a vision transformer-based pan-cancer foundation model trained with self-supervised learning to extract features from digitized H&E-stained slides. These features were integrated with clinical data to form a multi-modal AI test predicting cancer recurrence and death. The test was developed and evaluated using data from a total of 8,161 breast cancer patients across 15 cohorts originating from seven countries. Of these, 3,502 patients from five cohorts were used exclusively for evaluation, while the remaining patients were used for training. Our test accurately predicted our primary endpoint, disease-free interval, in the five external cohorts (C-index: 0.71 [0.68-0.75], HR: 3.63 [3.02-4.37, p<0.01]). In a direct comparison (N=858), the AI test was more accurate than Oncotype DX, the standard-of-care 21-gene assay, with a C-index of 0.67 [0.61-0.74] versus 0.61 [0.49-0.73], respectively. Additionally, the AI test added independent information to Oncotype DX in a multivariate analysis (HR: 3.11 [1.91-5.09, p<0.01)]). The test demonstrated robust accuracy across all major breast cancer subtypes, including TNBC (C-index: 0.71 [0.62-0.81], HR: 3.81 [2.35-6.17, p=0.02]), where no diagnostic tools are currently recommended by clinical guidelines. These results suggest that our AI test can improve accuracy, extend applicability to a wider range of patients, and enhance access to treatment selection tools.
AutoGLM: Autonomous Foundation Agents for GUIs
Oct 28, 2024



Abstract:We present AutoGLM, a new series in the ChatGLM family, designed to serve as foundation agents for autonomous control of digital devices through Graphical User Interfaces (GUIs). While foundation models excel at acquiring human knowledge, they often struggle with decision-making in dynamic real-world environments, limiting their progress toward artificial general intelligence. This limitation underscores the importance of developing foundation agents capable of learning through autonomous environmental interactions by reinforcing existing models. Focusing on Web Browser and Phone as representative GUI scenarios, we have developed AutoGLM as a practical foundation agent system for real-world GUI interactions. Our approach integrates a comprehensive suite of techniques and infrastructures to create deployable agent systems suitable for user delivery. Through this development, we have derived two key insights: First, the design of an appropriate "intermediate interface" for GUI control is crucial, enabling the separation of planning and grounding behaviors, which require distinct optimization for flexibility and accuracy respectively. Second, we have developed a novel progressive training framework that enables self-evolving online curriculum reinforcement learning for AutoGLM. Our evaluations demonstrate AutoGLM's effectiveness across multiple domains. For web browsing, AutoGLM achieves a 55.2% success rate on VAB-WebArena-Lite (improving to 59.1% with a second attempt) and 96.2% on OpenTable evaluation tasks. In Android device control, AutoGLM attains a 36.2% success rate on AndroidLab (VAB-Mobile) and 89.7% on common tasks in popular Chinese APPs.
Quantifying Impairment and Disease Severity Using AI Models Trained on Healthy Subjects
Nov 21, 2023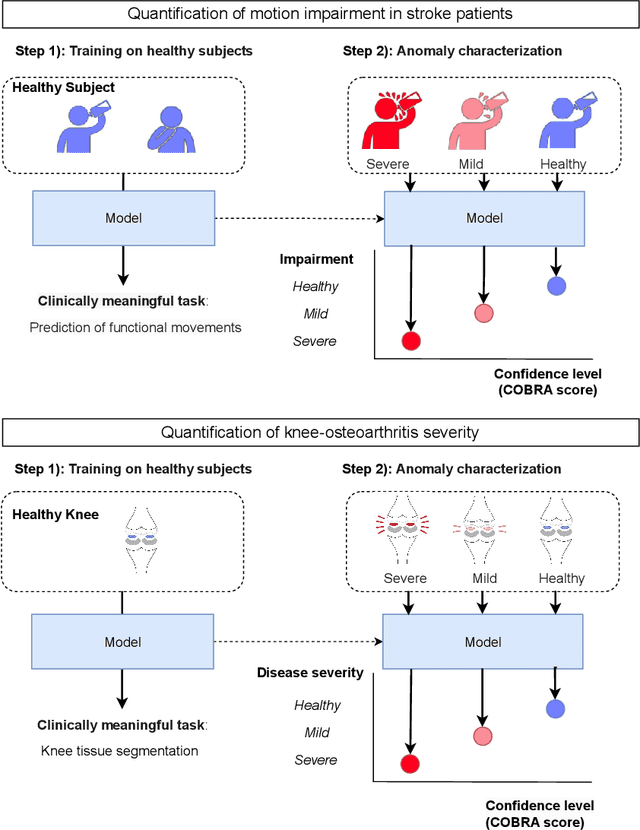
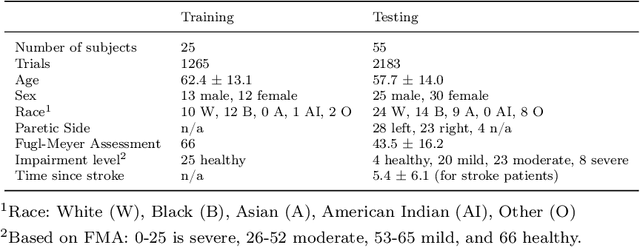
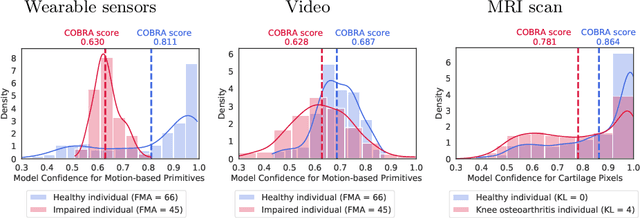
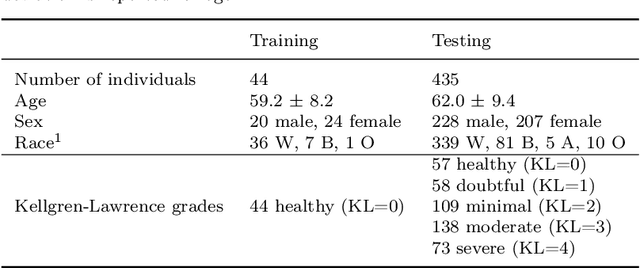
Abstract:Automatic assessment of impairment and disease severity is a key challenge in data-driven medicine. We propose a novel framework to address this challenge, which leverages AI models trained exclusively on healthy individuals. The COnfidence-Based chaRacterization of Anomalies (COBRA) score exploits the decrease in confidence of these models when presented with impaired or diseased patients to quantify their deviation from the healthy population. We applied the COBRA score to address a key limitation of current clinical evaluation of upper-body impairment in stroke patients. The gold-standard Fugl-Meyer Assessment (FMA) requires in-person administration by a trained assessor for 30-45 minutes, which restricts monitoring frequency and precludes physicians from adapting rehabilitation protocols to the progress of each patient. The COBRA score, computed automatically in under one minute, is shown to be strongly correlated with the FMA on an independent test cohort for two different data modalities: wearable sensors ($\rho = 0.845$, 95% CI [0.743,0.908]) and video ($\rho = 0.746$, 95% C.I [0.594, 0.847]). To demonstrate the generalizability of the approach to other conditions, the COBRA score was also applied to quantify severity of knee osteoarthritis from magnetic-resonance imaging scans, again achieving significant correlation with an independent clinical assessment ($\rho = 0.644$, 95% C.I [0.585,0.696]).
Controllable One-Shot Face Video Synthesis With Semantic Aware Prior
Apr 27, 2023Abstract:The one-shot talking-head synthesis task aims to animate a source image to another pose and expression, which is dictated by a driving frame. Recent methods rely on warping the appearance feature extracted from the source, by using motion fields estimated from the sparse keypoints, that are learned in an unsupervised manner. Due to their lightweight formulation, they are suitable for video conferencing with reduced bandwidth. However, based on our study, current methods suffer from two major limitations: 1) unsatisfactory generation quality in the case of large head poses and the existence of observable pose misalignment between the source and the first frame in driving videos. 2) fail to capture fine yet critical face motion details due to the lack of semantic understanding and appropriate face geometry regularization. To address these shortcomings, we propose a novel method that leverages the rich face prior information, the proposed model can generate face videos with improved semantic consistency (improve baseline by $7\%$ in average keypoint distance) and expression-preserving (outperform baseline by $15 \%$ in average emotion embedding distance) under equivalent bandwidth. Additionally, incorporating such prior information provides us with a convenient interface to achieve highly controllable generation in terms of both pose and expression.
Multiple Instance Learning via Iterative Self-Paced Supervised Contrastive Learning
Oct 17, 2022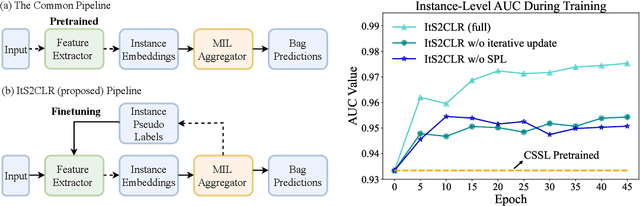
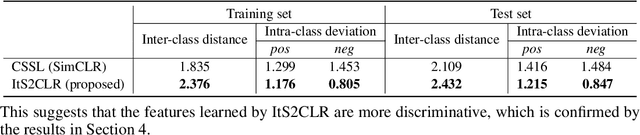
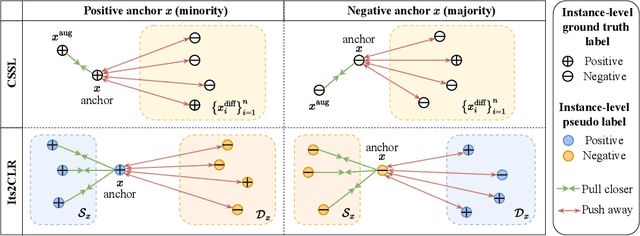

Abstract:Learning representations for individual instances when only bag-level labels are available is a fundamental challenge in multiple instance learning (MIL). Recent works have shown promising results using contrastive self-supervised learning (CSSL), which learns to push apart representations corresponding to two different randomly-selected instances. Unfortunately, in real-world applications such as medical image classification, there is often class imbalance, so randomly-selected instances mostly belong to the same majority class, which precludes CSSL from learning inter-class differences. To address this issue, we propose a novel framework, Iterative Self-paced Supervised Contrastive Learning for MIL Representations (ItS2CLR), which improves the learned representation by exploiting instance-level pseudo labels derived from the bag-level labels. The framework employs a novel self-paced sampling strategy to ensure the accuracy of pseudo labels. We evaluate ItS2CLR on three medical datasets, showing that it improves the quality of instance-level pseudo labels and representations, and outperforms existing MIL methods in terms of both bag and instance level accuracy.
Are All Losses Created Equal: A Neural Collapse Perspective
Oct 08, 2022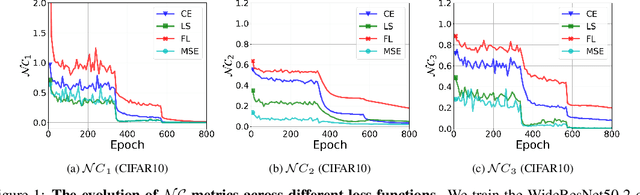
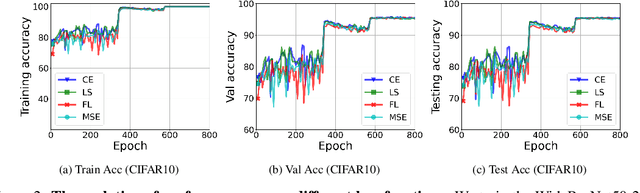
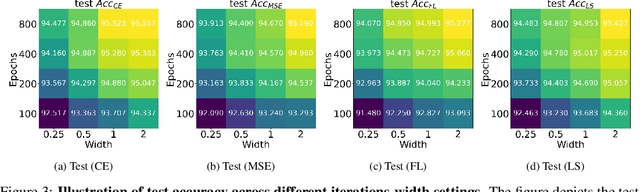
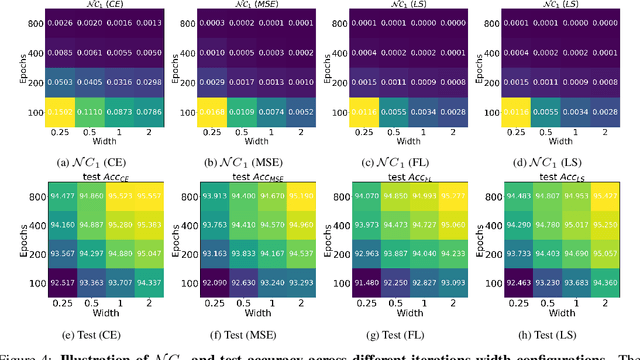
Abstract:While cross entropy (CE) is the most commonly used loss to train deep neural networks for classification tasks, many alternative losses have been developed to obtain better empirical performance. Among them, which one is the best to use is still a mystery, because there seem to be multiple factors affecting the answer, such as properties of the dataset, the choice of network architecture, and so on. This paper studies the choice of loss function by examining the last-layer features of deep networks, drawing inspiration from a recent line work showing that the global optimal solution of CE and mean-square-error (MSE) losses exhibits a Neural Collapse phenomenon. That is, for sufficiently large networks trained until convergence, (i) all features of the same class collapse to the corresponding class mean and (ii) the means associated with different classes are in a configuration where their pairwise distances are all equal and maximized. We extend such results and show through global solution and landscape analyses that a broad family of loss functions including commonly used label smoothing (LS) and focal loss (FL) exhibits Neural Collapse. Hence, all relevant losses(i.e., CE, LS, FL, MSE) produce equivalent features on training data. Based on the unconstrained feature model assumption, we provide either the global landscape analysis for LS loss or the local landscape analysis for FL loss and show that the (only!) global minimizers are neural collapse solutions, while all other critical points are strict saddles whose Hessian exhibit negative curvature directions either in the global scope for LS loss or in the local scope for FL loss near the optimal solution. The experiments further show that Neural Collapse features obtained from all relevant losses lead to largely identical performance on test data as well, provided that the network is sufficiently large and trained until convergence.
Sequence-to-Sequence Modeling for Action Identification at High Temporal Resolution
Nov 03, 2021
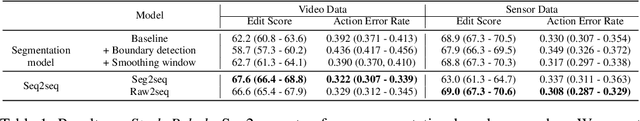
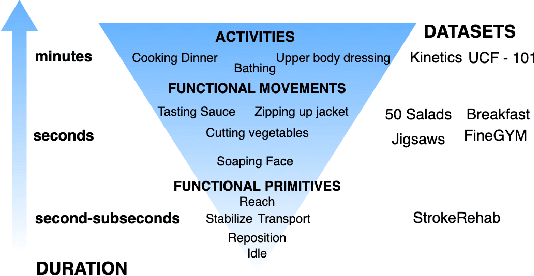
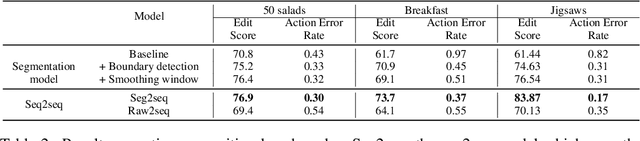
Abstract:Automatic action identification from video and kinematic data is an important machine learning problem with applications ranging from robotics to smart health. Most existing works focus on identifying coarse actions such as running, climbing, or cutting a vegetable, which have relatively long durations. This is an important limitation for applications that require the identification of subtle motions at high temporal resolution. For example, in stroke recovery, quantifying rehabilitation dose requires differentiating motions with sub-second durations. Our goal is to bridge this gap. To this end, we introduce a large-scale, multimodal dataset, StrokeRehab, as a new action-recognition benchmark that includes subtle short-duration actions labeled at a high temporal resolution. These short-duration actions are called functional primitives, and consist of reaches, transports, repositions, stabilizations, and idles. The dataset consists of high-quality Inertial Measurement Unit sensors and video data of 41 stroke-impaired patients performing activities of daily living like feeding, brushing teeth, etc. We show that current state-of-the-art models based on segmentation produce noisy predictions when applied to these data, which often leads to overcounting of actions. To address this, we propose a novel approach for high-resolution action identification, inspired by speech-recognition techniques, which is based on a sequence-to-sequence model that directly predicts the sequence of actions. This approach outperforms current state-of-the-art methods on the StrokeRehab dataset, as well as on the standard benchmark datasets 50Salads, Breakfast, and Jigsaws.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge