Jakub Chłędowski
3D-GMIC: an efficient deep neural network to find small objects in large 3D images
Oct 16, 2022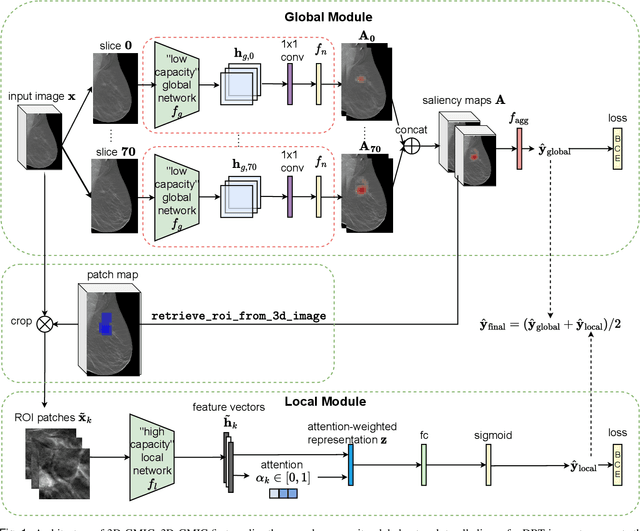
Abstract:3D imaging enables a more accurate diagnosis by providing spatial information about organ anatomy. However, using 3D images to train AI models is computationally challenging because they consist of tens or hundreds of times more pixels than their 2D counterparts. To train with high-resolution 3D images, convolutional neural networks typically resort to downsampling them or projecting them to two dimensions. In this work, we propose an effective alternative, a novel neural network architecture that enables computationally efficient classification of 3D medical images in their full resolution. Compared to off-the-shelf convolutional neural networks, 3D-GMIC uses 77.98%-90.05% less GPU memory and 91.23%-96.02% less computation. While our network is trained only with image-level labels, without segmentation labels, it explains its classification predictions by providing pixel-level saliency maps. On a dataset collected at NYU Langone Health, including 85,526 patients with full-field 2D mammography (FFDM), synthetic 2D mammography, and 3D mammography (DBT), our model, the 3D Globally-Aware Multiple Instance Classifier (3D-GMIC), achieves a breast-wise AUC of 0.831 (95% CI: 0.769-0.887) in classifying breasts with malignant findings using DBT images. As DBT and 2D mammography capture different information, averaging predictions on 2D and 3D mammography together leads to a diverse ensemble with an improved breast-wise AUC of 0.841 (95% CI: 0.768-0.895). Our model generalizes well to an external dataset from Duke University Hospital, achieving an image-wise AUC of 0.848 (95% CI: 0.798-0.896) in classifying DBT images with malignant findings.
Meta-repository of screening mammography classifiers
Aug 10, 2021
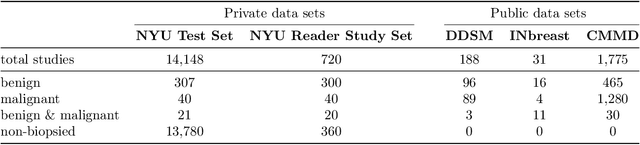
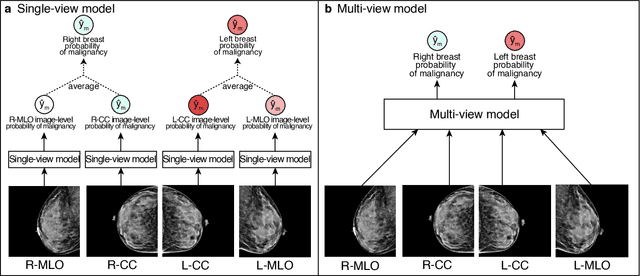
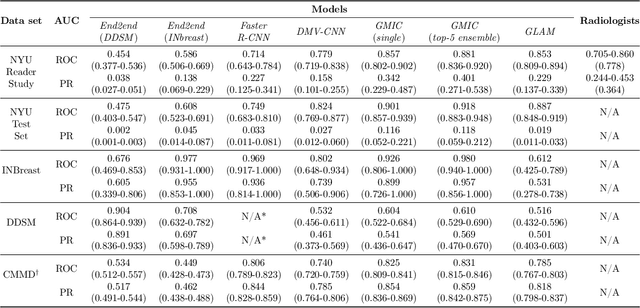
Abstract:Artificial intelligence (AI) is transforming medicine and showing promise in improving clinical diagnosis. In breast cancer screening, several recent studies show that AI has the potential to improve radiologists' accuracy, subsequently helping in early cancer diagnosis and reducing unnecessary workup. As the number of proposed models and their complexity grows, it is becoming increasingly difficult to re-implement them in order to reproduce the results and to compare different approaches. To enable reproducibility of research in this application area and to enable comparison between different methods, we release a meta-repository containing deep learning models for classification of screening mammograms. This meta-repository creates a framework that enables the evaluation of machine learning models on any private or public screening mammography data set. At its inception, our meta-repository contains five state-of-the-art models with open-source implementations and cross-platform compatibility. We compare their performance on five international data sets: two private New York University breast cancer screening data sets as well as three public (DDSM, INbreast and Chinese Mammography Database) data sets. Our framework has a flexible design that can be generalized to other medical image analysis tasks. The meta-repository is available at https://www.github.com/nyukat/mammography_metarepository.
Robust Learning-Augmented Caching: An Experimental Study
Jun 28, 2021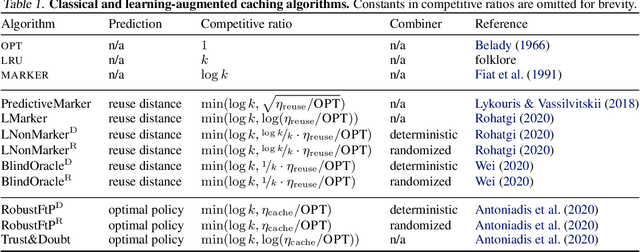
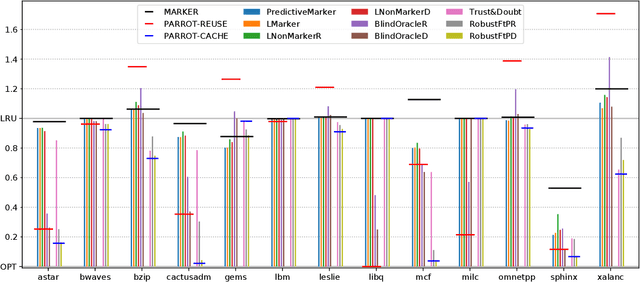
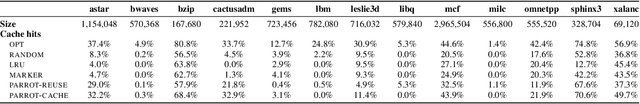
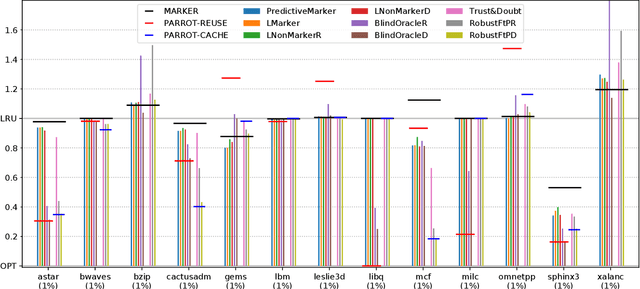
Abstract:Effective caching is crucial for the performance of modern-day computing systems. A key optimization problem arising in caching -- which item to evict to make room for a new item -- cannot be optimally solved without knowing the future. There are many classical approximation algorithms for this problem, but more recently researchers started to successfully apply machine learning to decide what to evict by discovering implicit input patterns and predicting the future. While machine learning typically does not provide any worst-case guarantees, the new field of learning-augmented algorithms proposes solutions that leverage classical online caching algorithms to make the machine-learned predictors robust. We are the first to comprehensively evaluate these learning-augmented algorithms on real-world caching datasets and state-of-the-art machine-learned predictors. We show that a straightforward method -- blindly following either a predictor or a classical robust algorithm, and switching whenever one becomes worse than the other -- has only a low overhead over a well-performing predictor, while competing with classical methods when the coupled predictor fails, thus providing a cheap worst-case insurance.
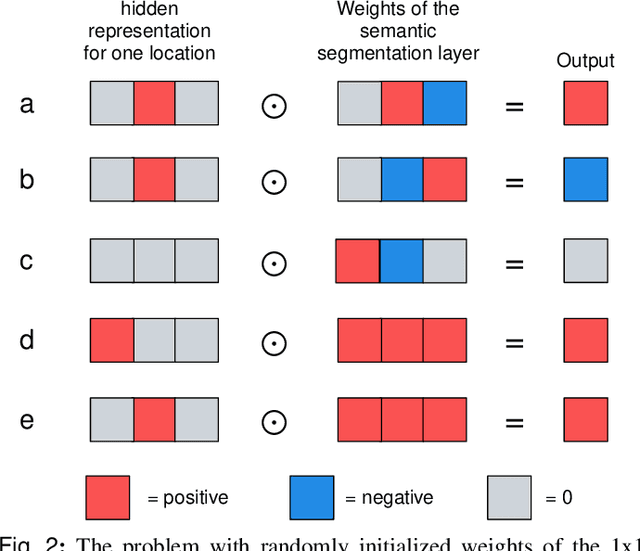
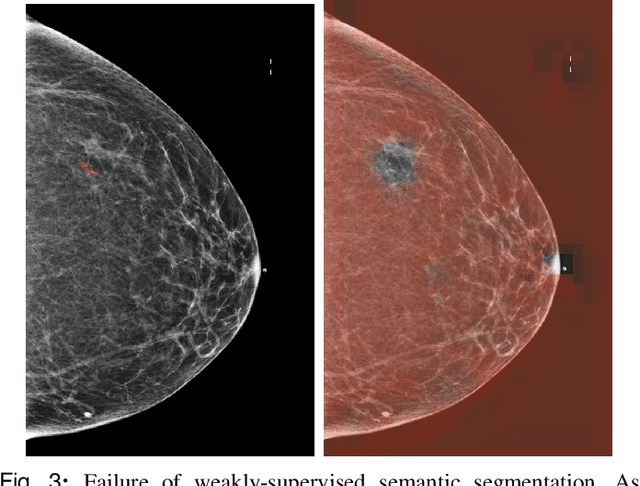
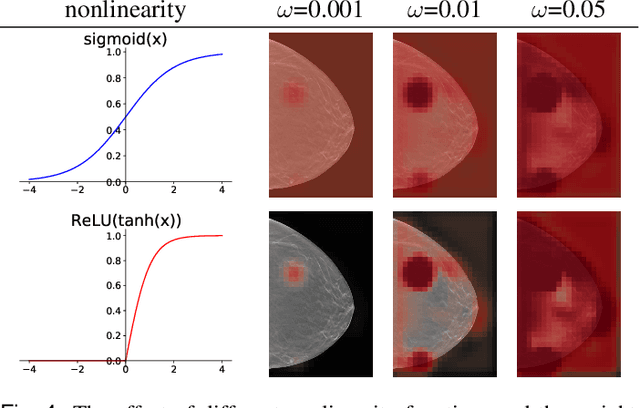
Weakly-supervised High-resolution Segmentation of Mammography Images for Breast Cancer Diagnosis
Jun 15, 2021



Abstract:In the last few years, deep learning classifiers have shown promising results in image-based medical diagnosis. However, interpreting the outputs of these models remains a challenge. In cancer diagnosis, interpretability can be achieved by localizing the region of the input image responsible for the output, i.e. the location of a lesion. Alternatively, segmentation or detection models can be trained with pixel-wise annotations indicating the locations of malignant lesions. Unfortunately, acquiring such labels is labor-intensive and requires medical expertise. To overcome this difficulty, weakly-supervised localization can be utilized. These methods allow neural network classifiers to output saliency maps highlighting the regions of the input most relevant to the classification task (e.g. malignant lesions in mammograms) using only image-level labels (e.g. whether the patient has cancer or not) during training. When applied to high-resolution images, existing methods produce low-resolution saliency maps. This is problematic in applications in which suspicious lesions are small in relation to the image size. In this work, we introduce a novel neural network architecture to perform weakly-supervised segmentation of high-resolution images. The proposed model selects regions of interest via coarse-level localization, and then performs fine-grained segmentation of those regions. We apply this model to breast cancer diagnosis with screening mammography, and validate it on a large clinically-realistic dataset. Measured by Dice similarity score, our approach outperforms existing methods by a large margin in terms of localization performance of benign and malignant lesions, relatively improving the performance by 39.6% and 20.0%, respectively. Code and the weights of some of the models are available at https://github.com/nyukat/GLAM
From Dataset Recycling to Multi-Property Extraction and Beyond
Nov 06, 2020



Abstract:This paper investigates various Transformer architectures on the WikiReading Information Extraction and Machine Reading Comprehension dataset. The proposed dual-source model outperforms the current state-of-the-art by a large margin. Next, we introduce WikiReading Recycled-a newly developed public dataset and the task of multiple property extraction. It uses the same data as WikiReading but does not inherit its predecessor's identified disadvantages. In addition, we provide a human-annotated test set with diagnostic subsets for a detailed analysis of model performance.
On the Multi-Property Extraction and Beyond
Jun 15, 2020
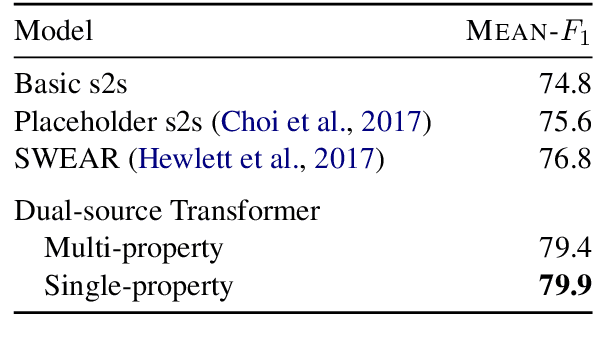
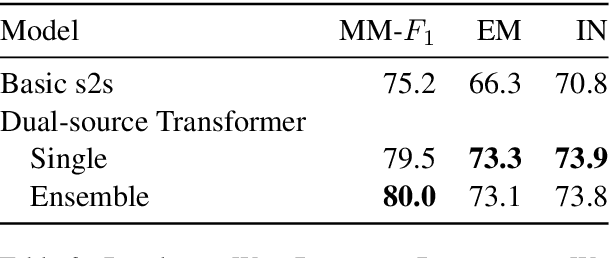
Abstract:In this paper, we investigate the Dual-source Transformer architecture on the WikiReading information extraction and machine reading comprehension dataset. The proposed model outperforms the current state-of-the-art by a large margin. Next, we introduce WikiReading Recycled - a newly developed public dataset, supporting the task of multiple property extraction. It keeps the spirit of the original WikiReading but does not inherit the identified disadvantages of its predecessor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge