3D-GMIC: an efficient deep neural network to find small objects in large 3D images
Paper and Code
Oct 16, 2022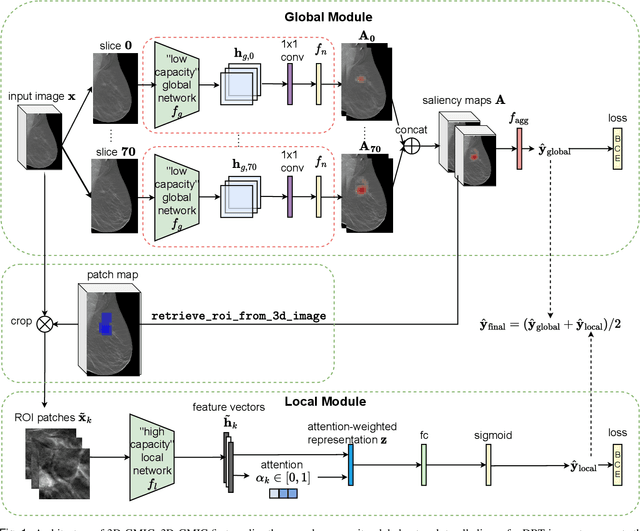
3D imaging enables a more accurate diagnosis by providing spatial information about organ anatomy. However, using 3D images to train AI models is computationally challenging because they consist of tens or hundreds of times more pixels than their 2D counterparts. To train with high-resolution 3D images, convolutional neural networks typically resort to downsampling them or projecting them to two dimensions. In this work, we propose an effective alternative, a novel neural network architecture that enables computationally efficient classification of 3D medical images in their full resolution. Compared to off-the-shelf convolutional neural networks, 3D-GMIC uses 77.98%-90.05% less GPU memory and 91.23%-96.02% less computation. While our network is trained only with image-level labels, without segmentation labels, it explains its classification predictions by providing pixel-level saliency maps. On a dataset collected at NYU Langone Health, including 85,526 patients with full-field 2D mammography (FFDM), synthetic 2D mammography, and 3D mammography (DBT), our model, the 3D Globally-Aware Multiple Instance Classifier (3D-GMIC), achieves a breast-wise AUC of 0.831 (95% CI: 0.769-0.887) in classifying breasts with malignant findings using DBT images. As DBT and 2D mammography capture different information, averaging predictions on 2D and 3D mammography together leads to a diverse ensemble with an improved breast-wise AUC of 0.841 (95% CI: 0.768-0.895). Our model generalizes well to an external dataset from Duke University Hospital, achieving an image-wise AUC of 0.848 (95% CI: 0.798-0.896) in classifying DBT images with malignant findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge