Farah E. Shamout
MedErrBench: A Fine-Grained Multilingual Benchmark for Medical Error Detection and Correction with Clinical Expert Annotations
Feb 05, 2026Abstract:Inaccuracies in existing or generated clinical text may lead to serious adverse consequences, especially if it is a misdiagnosis or incorrect treatment suggestion. With Large Language Models (LLMs) increasingly being used across diverse healthcare applications, comprehensive evaluation through dedicated benchmarks is crucial. However, such datasets remain scarce, especially across diverse languages and contexts. In this paper, we introduce MedErrBench, the first multilingual benchmark for error detection, localization, and correction, developed under the guidance of experienced clinicians. Based on an expanded taxonomy of ten common error types, MedErrBench covers English, Arabic and Chinese, with natural clinical cases annotated and reviewed by domain experts. We assessed the performance of a range of general-purpose, language-specific, and medical-domain language models across all three tasks. Our results reveal notable performance gaps, particularly in non-English settings, highlighting the need for clinically grounded, language-aware systems. By making MedErrBench and our evaluation protocols publicly-available, we aim to advance multilingual clinical NLP to promote safer and more equitable AI-based healthcare globally. The dataset is available in the supplementary material. An anonymized version of the dataset is available at: https://github.com/congboma/MedErrBench.
Cross-Lingual Empirical Evaluation of Large Language Models for Arabic Medical Tasks
Feb 05, 2026Abstract:In recent years, Large Language Models (LLMs) have become widely used in medical applications, such as clinical decision support, medical education, and medical question answering. Yet, these models are often English-centric, limiting their robustness and reliability for linguistically diverse communities. Recent work has highlighted discrepancies in performance in low-resource languages for various medical tasks, but the underlying causes remain poorly understood. In this study, we conduct a cross-lingual empirical analysis of LLM performance on Arabic and English medical question and answering. Our findings reveal a persistent language-driven performance gap that intensifies with increasing task complexity. Tokenization analysis exposes structural fragmentation in Arabic medical text, while reliability analysis suggests that model-reported confidence and explanations exhibit limited correlation with correctness. Together, these findings underscore the need for language-aware design and evaluation strategies in LLMs for medical tasks.
MedAraBench: Large-Scale Arabic Medical Question Answering Dataset and Benchmark
Feb 02, 2026Abstract:Arabic remains one of the most underrepresented languages in natural language processing research, particularly in medical applications, due to the limited availability of open-source data and benchmarks. The lack of resources hinders efforts to evaluate and advance the multilingual capabilities of Large Language Models (LLMs). In this paper, we introduce MedAraBench, a large-scale dataset consisting of Arabic multiple-choice question-answer pairs across various medical specialties. We constructed the dataset by manually digitizing a large repository of academic materials created by medical professionals in the Arabic-speaking region. We then conducted extensive preprocessing and split the dataset into training and test sets to support future research efforts in the area. To assess the quality of the data, we adopted two frameworks, namely expert human evaluation and LLM-as-a-judge. Our dataset is diverse and of high quality, spanning 19 specialties and five difficulty levels. For benchmarking purposes, we assessed the performance of eight state-of-the-art open-source and proprietary models, such as GPT-5, Gemini 2.0 Flash, and Claude 4-Sonnet. Our findings highlight the need for further domain-specific enhancements. We release the dataset and evaluation scripts to broaden the diversity of medical data benchmarks, expand the scope of evaluation suites for LLMs, and enhance the multilingual capabilities of models for deployment in clinical settings.
MedArabiQ: Benchmarking Large Language Models on Arabic Medical Tasks
May 06, 2025



Abstract:Large Language Models (LLMs) have demonstrated significant promise for various applications in healthcare. However, their efficacy in the Arabic medical domain remains unexplored due to the lack of high-quality domain-specific datasets and benchmarks. This study introduces MedArabiQ, a novel benchmark dataset consisting of seven Arabic medical tasks, covering multiple specialties and including multiple choice questions, fill-in-the-blank, and patient-doctor question answering. We first constructed the dataset using past medical exams and publicly available datasets. We then introduced different modifications to evaluate various LLM capabilities, including bias mitigation. We conducted an extensive evaluation with five state-of-the-art open-source and proprietary LLMs, including GPT-4o, Claude 3.5-Sonnet, and Gemini 1.5. Our findings highlight the need for the creation of new high-quality benchmarks that span different languages to ensure fair deployment and scalability of LLMs in healthcare. By establishing this benchmark and releasing the dataset, we provide a foundation for future research aimed at evaluating and enhancing the multilingual capabilities of LLMs for the equitable use of generative AI in healthcare.
Multimodal Deep Learning for Stroke Prediction and Detection using Retinal Imaging and Clinical Data
May 05, 2025
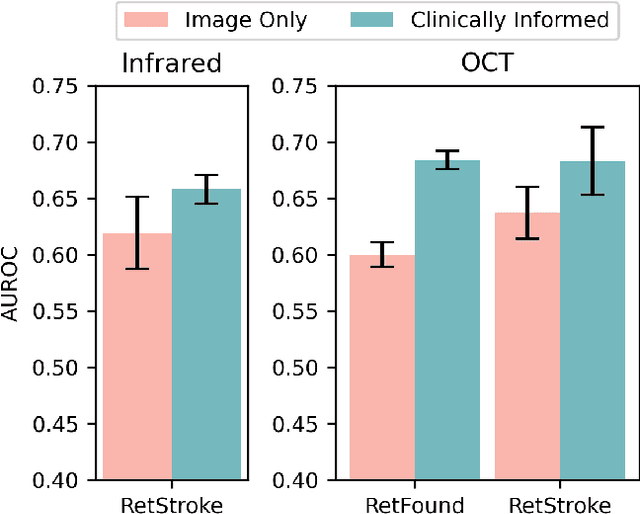
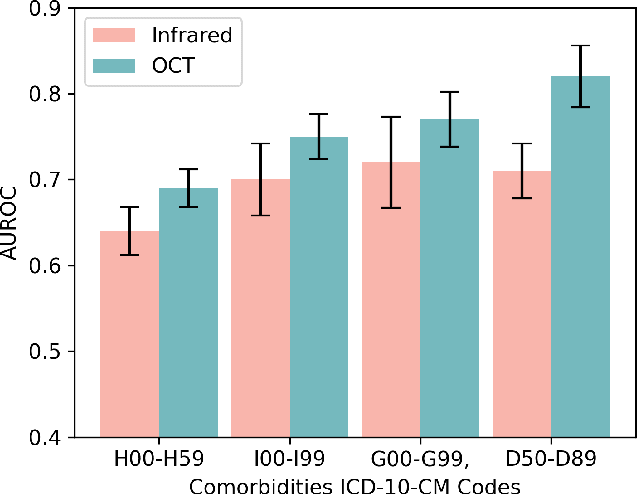
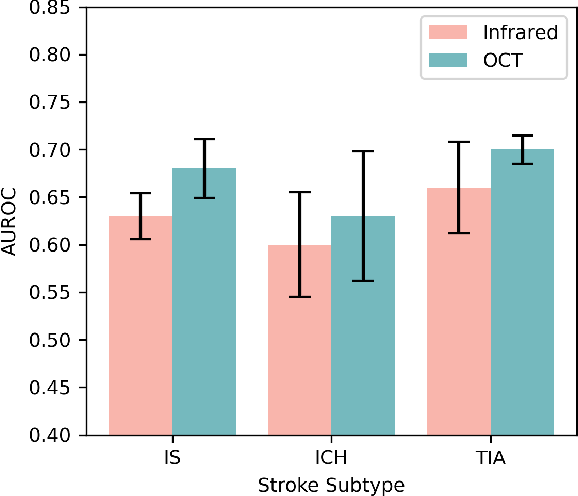
Abstract:Stroke is a major public health problem, affecting millions worldwide. Deep learning has recently demonstrated promise for enhancing the diagnosis and risk prediction of stroke. However, existing methods rely on costly medical imaging modalities, such as computed tomography. Recent studies suggest that retinal imaging could offer a cost-effective alternative for cerebrovascular health assessment due to the shared clinical pathways between the retina and the brain. Hence, this study explores the impact of leveraging retinal images and clinical data for stroke detection and risk prediction. We propose a multimodal deep neural network that processes Optical Coherence Tomography (OCT) and infrared reflectance retinal scans, combined with clinical data, such as demographics, vital signs, and diagnosis codes. We pretrained our model using a self-supervised learning framework using a real-world dataset consisting of $37$ k scans, and then fine-tuned and evaluated the model using a smaller labeled subset. Our empirical findings establish the predictive ability of the considered modalities in detecting lasting effects in the retina associated with acute stroke and forecasting future risk within a specific time horizon. The experimental results demonstrate the effectiveness of our proposed framework by achieving $5$\% AUROC improvement as compared to the unimodal image-only baseline, and $8$\% improvement compared to an existing state-of-the-art foundation model. In conclusion, our study highlights the potential of retinal imaging in identifying high-risk patients and improving long-term outcomes.
Uncertainty Quantification for Machine Learning in Healthcare: A Survey
May 04, 2025

Abstract:Uncertainty Quantification (UQ) is pivotal in enhancing the robustness, reliability, and interpretability of Machine Learning (ML) systems for healthcare, optimizing resources and improving patient care. Despite the emergence of ML-based clinical decision support tools, the lack of principled quantification of uncertainty in ML models remains a major challenge. Current reviews have a narrow focus on analyzing the state-of-the-art UQ in specific healthcare domains without systematically evaluating method efficacy across different stages of model development, and despite a growing body of research, its implementation in healthcare applications remains limited. Therefore, in this survey, we provide a comprehensive analysis of current UQ in healthcare, offering an informed framework that highlights how different methods can be integrated into each stage of the ML pipeline including data processing, training and evaluation. We also highlight the most popular methods used in healthcare and novel approaches from other domains that hold potential for future adoption in the medical context. We expect this study will provide a clear overview of the challenges and opportunities of implementing UQ in the ML pipeline for healthcare, guiding researchers and practitioners in selecting suitable techniques to enhance the reliability, safety and trust from patients and clinicians on ML-driven healthcare solutions.
MIND: Modality-Informed Knowledge Distillation Framework for Multimodal Clinical Prediction Tasks
Feb 03, 2025Abstract:Multimodal fusion leverages information across modalities to learn better feature representations with the goal of improving performance in fusion-based tasks. However, multimodal datasets, especially in medical settings, are typically smaller than their unimodal counterparts, which can impede the performance of multimodal models. Additionally, the increase in the number of modalities is often associated with an overall increase in the size of the multimodal network, which may be undesirable in medical use cases. Utilizing smaller unimodal encoders may lead to sub-optimal performance, particularly when dealing with high-dimensional clinical data. In this paper, we propose the Modality-INformed knowledge Distillation (MIND) framework, a multimodal model compression approach based on knowledge distillation that transfers knowledge from ensembles of pre-trained deep neural networks of varying sizes into a smaller multimodal student. The teacher models consist of unimodal networks, allowing the student to learn from diverse representations. MIND employs multi-head joint fusion models, as opposed to single-head models, enabling the use of unimodal encoders in the case of unimodal samples without requiring imputation or masking of absent modalities. As a result, MIND generates an optimized multimodal model, enhancing both multimodal and unimodal representations. It can also be leveraged to balance multimodal learning during training. We evaluate MIND on binary and multilabel clinical prediction tasks using time series data and chest X-ray images. Additionally, we assess the generalizability of the MIND framework on three non-medical multimodal multiclass datasets. Experimental results demonstrate that MIND enhances the performance of the smaller multimodal network across all five tasks, as well as various fusion methods and multimodal architectures, compared to state-of-the-art baselines.
* Published in Transactions on Machine Learning Research (01/2025), https://openreview.net/forum?id=BhOJreYmur¬eId=ymnAhncuez
The Role of Functional Muscle Networks in Improving Hand Gesture Perception for Human-Machine Interfaces
Aug 05, 2024Abstract:Developing accurate hand gesture perception models is critical for various robotic applications, enabling effective communication between humans and machines and directly impacting neurorobotics and interactive robots. Recently, surface electromyography (sEMG) has been explored for its rich informational context and accessibility when combined with advanced machine learning approaches and wearable systems. The literature presents numerous approaches to boost performance while ensuring robustness for neurorobots using sEMG, often resulting in models requiring high processing power, large datasets, and less scalable solutions. This paper addresses this challenge by proposing the decoding of muscle synchronization rather than individual muscle activation. We study coherence-based functional muscle networks as the core of our perception model, proposing that functional synchronization between muscles and the graph-based network of muscle connectivity encode contextual information about intended hand gestures. This can be decoded using shallow machine learning approaches without the need for deep temporal networks. Our technique could impact myoelectric control of neurorobots by reducing computational burdens and enhancing efficiency. The approach is benchmarked on the Ninapro database, which contains 12 EMG signals from 40 subjects performing 17 hand gestures. It achieves an accuracy of 85.1%, demonstrating improved performance compared to existing methods while requiring much less computational power. The results support the hypothesis that a coherence-based functional muscle network encodes critical information related to gesture execution, significantly enhancing hand gesture perception with potential applications for neurorobotic systems and interactive machines.
Multi-modal Masked Siamese Network Improves Chest X-Ray Representation Learning
Jul 05, 2024Abstract:Self-supervised learning methods for medical images primarily rely on the imaging modality during pretraining. While such approaches deliver promising results, they do not leverage associated patient or scan information collected within Electronic Health Records (EHR). Here, we propose to incorporate EHR data during self-supervised pretraining with a Masked Siamese Network (MSN) to enhance the quality of chest X-ray representations. We investigate three types of EHR data, including demographic, scan metadata, and inpatient stay information. We evaluate our approach on three publicly available chest X-ray datasets, MIMIC-CXR, CheXpert, and NIH-14, using two vision transformer (ViT) backbones, specifically ViT-Tiny and ViT-Small. In assessing the quality of the representations via linear evaluation, our proposed method demonstrates significant improvement compared to vanilla MSN and state-of-the-art self-supervised learning baselines. Our work highlights the potential of EHR-enhanced self-supervised pre-training for medical imaging. The code is publicly available at: https://github.com/nyuad-cai/CXR-EHR-MSN
Informative Priors Improve the Reliability of Multimodal Clinical Data Classification
Nov 17, 2023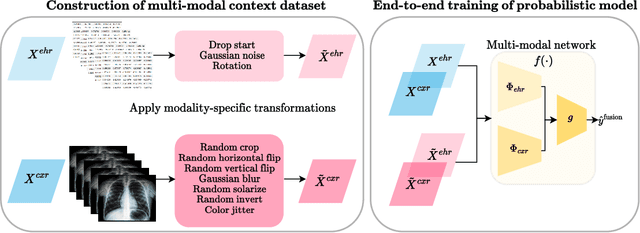

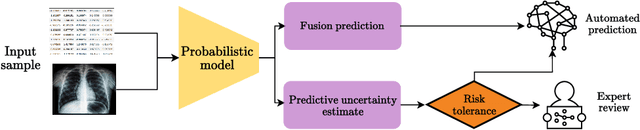
Abstract:Machine learning-aided clinical decision support has the potential to significantly improve patient care. However, existing efforts in this domain for principled quantification of uncertainty have largely been limited to applications of ad-hoc solutions that do not consistently improve reliability. In this work, we consider stochastic neural networks and design a tailor-made multimodal data-driven (M2D2) prior distribution over network parameters. We use simple and scalable Gaussian mean-field variational inference to train a Bayesian neural network using the M2D2 prior. We train and evaluate the proposed approach using clinical time-series data in MIMIC-IV and corresponding chest X-ray images in MIMIC-CXR for the classification of acute care conditions. Our empirical results show that the proposed method produces a more reliable predictive model compared to deterministic and Bayesian neural network baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge