Saeed Shurrab
Multimodal Deep Learning for Stroke Prediction and Detection using Retinal Imaging and Clinical Data
May 05, 2025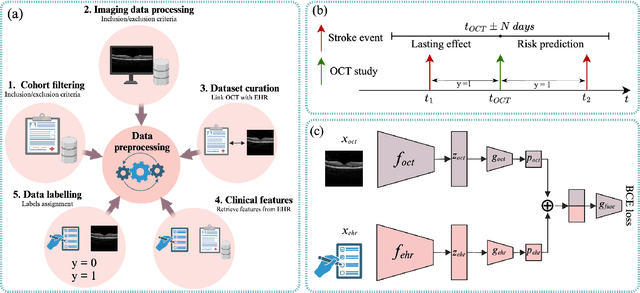
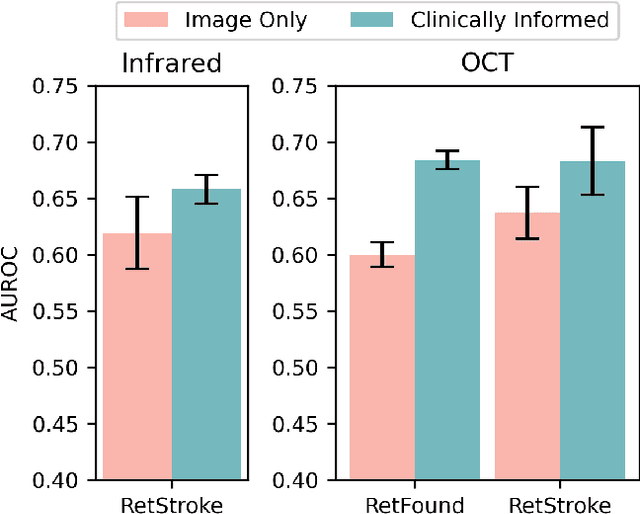
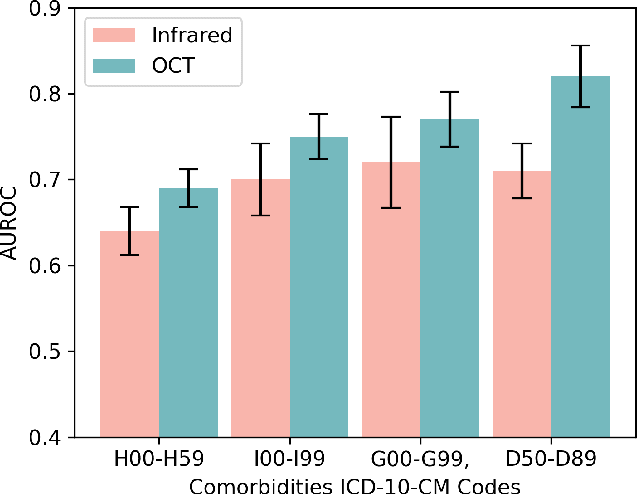
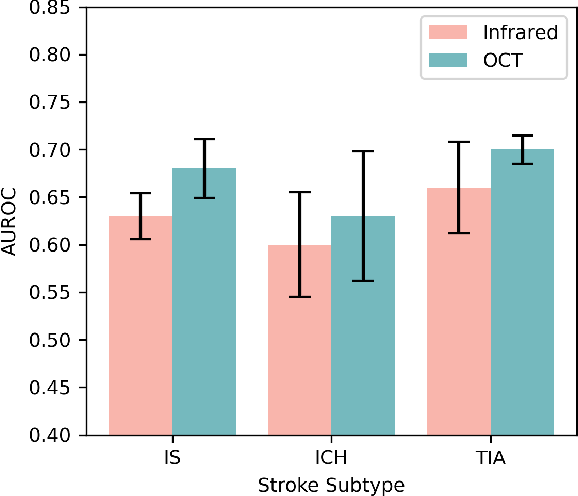
Abstract:Stroke is a major public health problem, affecting millions worldwide. Deep learning has recently demonstrated promise for enhancing the diagnosis and risk prediction of stroke. However, existing methods rely on costly medical imaging modalities, such as computed tomography. Recent studies suggest that retinal imaging could offer a cost-effective alternative for cerebrovascular health assessment due to the shared clinical pathways between the retina and the brain. Hence, this study explores the impact of leveraging retinal images and clinical data for stroke detection and risk prediction. We propose a multimodal deep neural network that processes Optical Coherence Tomography (OCT) and infrared reflectance retinal scans, combined with clinical data, such as demographics, vital signs, and diagnosis codes. We pretrained our model using a self-supervised learning framework using a real-world dataset consisting of $37$ k scans, and then fine-tuned and evaluated the model using a smaller labeled subset. Our empirical findings establish the predictive ability of the considered modalities in detecting lasting effects in the retina associated with acute stroke and forecasting future risk within a specific time horizon. The experimental results demonstrate the effectiveness of our proposed framework by achieving $5$\% AUROC improvement as compared to the unimodal image-only baseline, and $8$\% improvement compared to an existing state-of-the-art foundation model. In conclusion, our study highlights the potential of retinal imaging in identifying high-risk patients and improving long-term outcomes.
Multi-modal Masked Siamese Network Improves Chest X-Ray Representation Learning
Jul 05, 2024Abstract:Self-supervised learning methods for medical images primarily rely on the imaging modality during pretraining. While such approaches deliver promising results, they do not leverage associated patient or scan information collected within Electronic Health Records (EHR). Here, we propose to incorporate EHR data during self-supervised pretraining with a Masked Siamese Network (MSN) to enhance the quality of chest X-ray representations. We investigate three types of EHR data, including demographic, scan metadata, and inpatient stay information. We evaluate our approach on three publicly available chest X-ray datasets, MIMIC-CXR, CheXpert, and NIH-14, using two vision transformer (ViT) backbones, specifically ViT-Tiny and ViT-Small. In assessing the quality of the representations via linear evaluation, our proposed method demonstrates significant improvement compared to vanilla MSN and state-of-the-art self-supervised learning baselines. Our work highlights the potential of EHR-enhanced self-supervised pre-training for medical imaging. The code is publicly available at: https://github.com/nyuad-cai/CXR-EHR-MSN
Self-supervised learning methods and applications in medical imaging analysis: A survey
Sep 25, 2021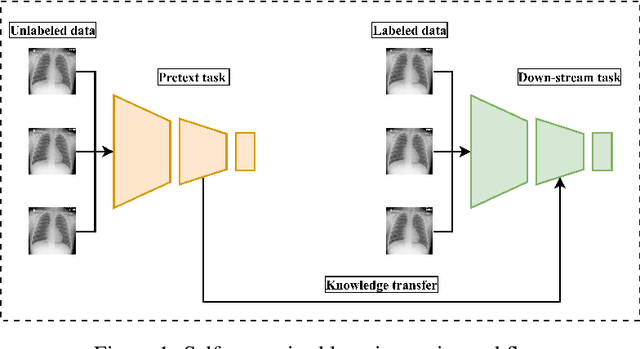
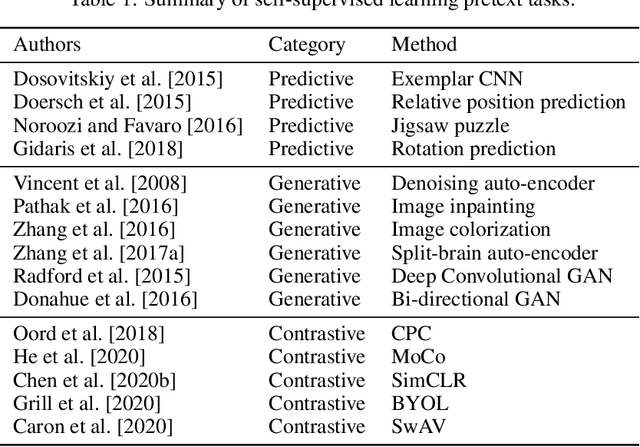

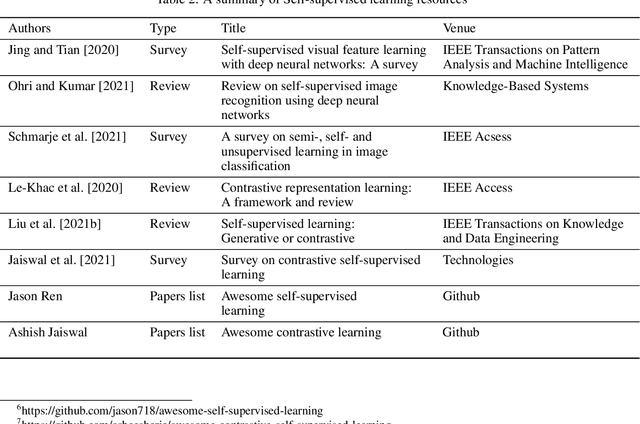
Abstract:The availability of high quality annotated medical imaging datasets is a major problem that collides with machine learning applications in the field of medical imaging analysis and impedes its advancement. Self-supervised learning is a recent training paradigm that enables learning robust representations without the need for human annotation which can be considered as an effective solution for the scarcity in annotated medical data. This article reviews the state-of-the-art research directions in self-supervised learning approaches for image data with concentration on their applications in the field of medical imaging analysis. The article covers a set of the most recent self-supervised learning methods from the computer vision field as they are applicable to the medical imaging analysis and categorize them as predictive, generative and contrastive approaches. Moreover, the article covers (40) of the most recent researches in the field of self-supervised learning in medical imaging analysis aiming at shedding the light on the recent innovation in the field. Ultimately, the article concludes with possible future research directions in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge