Sumit Chopra
DIMCIM: A Quantitative Evaluation Framework for Default-mode Diversity and Generalization in Text-to-Image Generative Models
Jun 05, 2025Abstract:Recent advances in text-to-image (T2I) models have achieved impressive quality and consistency. However, this has come at the cost of representation diversity. While automatic evaluation methods exist for benchmarking model diversity, they either require reference image datasets or lack specificity about the kind of diversity measured, limiting their adaptability and interpretability. To address this gap, we introduce the Does-it/Can-it framework, DIM-CIM, a reference-free measurement of default-mode diversity ("Does" the model generate images with expected attributes?) and generalization capacity ("Can" the model generate diverse attributes for a particular concept?). We construct the COCO-DIMCIM benchmark, which is seeded with COCO concepts and captions and augmented by a large language model. With COCO-DIMCIM, we find that widely-used models improve in generalization at the cost of default-mode diversity when scaling from 1.5B to 8.1B parameters. DIMCIM also identifies fine-grained failure cases, such as attributes that are generated with generic prompts but are rarely generated when explicitly requested. Finally, we use DIMCIM to evaluate the training data of a T2I model and observe a correlation of 0.85 between diversity in training images and default-mode diversity. Our work provides a flexible and interpretable framework for assessing T2I model diversity and generalization, enabling a more comprehensive understanding of model performance.
A Trust-Guided Approach to MR Image Reconstruction with Side Information
Jan 06, 2025



Abstract:Reducing MRI scan times can improve patient care and lower healthcare costs. Many acceleration methods are designed to reconstruct diagnostic-quality images from limited sets of acquired $\textit{k}$-space data. This task can be framed as a linear inverse problem (LIP), where, as a result of undersampling, the forward operator may become rank-deficient or exhibit small singular values. This results in ambiguities in reconstruction, in which multiple generally incorrect or non-diagnostic images can map to the same acquired data. To address such ambiguities, it is crucial to incorporate prior knowledge, for example in the form of regularization. Another form of prior knowledge less commonly used in medical imaging is contextual side information garnered from other sources than the current acquisition. Here, we propose the $\textbf{T}$rust-$\textbf{G}$uided $\textbf{V}$ariational $\textbf{N}$etwork $\textbf{(TGVN)}$, a novel end-to-end deep learning framework that effectively integrates side information into LIPs. TGVN eliminates undesirable solutions from the ambiguous space of the forward operator while remaining faithful to the acquired data. We demonstrate its effectiveness in multi-coil, multi-contrast MR image reconstruction, where incomplete or low-quality measurements from one contrast are used as side information to reconstruct high-quality images of another contrast from heavily under-sampled data. Our method is robust across different contrasts, anatomies, and field strengths. Compared to baselines that also utilize side information, TGVN achieves superior image quality at challenging under-sampling levels, drastically speeding up acquisition while minimizing hallucinations. Our approach is also versatile enough to incorporate many different types of side information (including previous scans or even text) into any LIP.
HIST-AID: Leveraging Historical Patient Reports for Enhanced Multi-Modal Automatic Diagnosis
Nov 16, 2024Abstract:Chest X-ray imaging is a widely accessible and non-invasive diagnostic tool for detecting thoracic abnormalities. While numerous AI models assist radiologists in interpreting these images, most overlook patients' historical data. To bridge this gap, we introduce Temporal MIMIC dataset, which integrates five years of patient history, including radiographic scans and reports from MIMIC-CXR and MIMIC-IV, encompassing 12,221 patients and thirteen pathologies. Building on this, we present HIST-AID, a framework that enhances automatic diagnostic accuracy using historical reports. HIST-AID emulates the radiologist's comprehensive approach, leveraging historical data to improve diagnostic accuracy. Our experiments demonstrate significant improvements, with AUROC increasing by 6.56% and AUPRC by 9.51% compared to models that rely solely on radiographic scans. These gains were consistently observed across diverse demographic groups, including variations in gender, age, and racial categories. We show that while recent data boost performance, older data may reduce accuracy due to changes in patient conditions. Our work paves the potential of incorporating historical data for more reliable automatic diagnosis, providing critical support for clinical decision-making.
Fine-Tuning In-House Large Language Models to Infer Differential Diagnosis from Radiology Reports
Oct 11, 2024



Abstract:Radiology reports summarize key findings and differential diagnoses derived from medical imaging examinations. The extraction of differential diagnoses is crucial for downstream tasks, including patient management and treatment planning. However, the unstructured nature of these reports, characterized by diverse linguistic styles and inconsistent formatting, presents significant challenges. Although proprietary large language models (LLMs) such as GPT-4 can effectively retrieve clinical information, their use is limited in practice by high costs and concerns over the privacy of protected health information (PHI). This study introduces a pipeline for developing in-house LLMs tailored to identify differential diagnoses from radiology reports. We first utilize GPT-4 to create 31,056 labeled reports, then fine-tune open source LLM using this dataset. Evaluated on a set of 1,067 reports annotated by clinicians, the proposed model achieves an average F1 score of 92.1\%, which is on par with GPT-4 (90.8\%). Through this study, we provide a methodology for constructing in-house LLMs that: match the performance of GPT, reduce dependence on expensive proprietary models, and enhance the privacy and security of PHI.
BURExtract-Llama: An LLM for Clinical Concept Extraction in Breast Ultrasound Reports
Aug 21, 2024



Abstract:Breast ultrasound is essential for detecting and diagnosing abnormalities, with radiology reports summarizing key findings like lesion characteristics and malignancy assessments. Extracting this critical information is challenging due to the unstructured nature of these reports, with varied linguistic styles and inconsistent formatting. While proprietary LLMs like GPT-4 are effective, they are costly and raise privacy concerns when handling protected health information. This study presents a pipeline for developing an in-house LLM to extract clinical information from radiology reports. We first use GPT-4 to create a small labeled dataset, then fine-tune a Llama3-8B model on it. Evaluated on clinician-annotated reports, our model achieves an average F1 score of 84.6%, which is on par with GPT-4. Our findings demonstrate the feasibility of developing an in-house LLM that not only matches GPT-4's performance but also offers cost reductions and enhanced data privacy.
A training regime to learn unified representations from complementary breast imaging modalities
Aug 16, 2024



Abstract:Full Field Digital Mammograms (FFDMs) and Digital Breast Tomosynthesis (DBT) are the two most widely used imaging modalities for breast cancer screening. Although DBT has increased cancer detection compared to FFDM, its widespread adoption in clinical practice has been slowed by increased interpretation times and a perceived decrease in the conspicuity of specific lesion types. Specifically, the non-inferiority of DBT for microcalcifications remains under debate. Due to concerns about the decrease in visual acuity, combined DBT-FFDM acquisitions remain popular, leading to overall increased exam times and radiation dosage. Enabling DBT to provide diagnostic information present in both FFDM and DBT would reduce reliance on FFDM, resulting in a reduction in both quantities. We propose a machine learning methodology that learns high-level representations leveraging the complementary diagnostic signal from both DBT and FFDM. Experiments on a large-scale data set validate our claims and show that our representations enable more accurate breast lesion detection than any DBT- or FFDM-based model.
Adaptive Sampling of k-Space in Magnetic Resonance for Rapid Pathology Prediction
Jun 06, 2024



Abstract:Magnetic Resonance (MR) imaging, despite its proven diagnostic utility, remains an inaccessible imaging modality for disease surveillance at the population level. A major factor rendering MR inaccessible is lengthy scan times. An MR scanner collects measurements associated with the underlying anatomy in the Fourier space, also known as the k-space. Creating a high-fidelity image requires collecting large quantities of such measurements, increasing the scan time. Traditionally to accelerate an MR scan, image reconstruction from under-sampled k-space data is the method of choice. However, recent works show the feasibility of bypassing image reconstruction and directly learning to detect disease directly from a sparser learned subset of the k-space measurements. In this work, we propose Adaptive Sampling for MR (ASMR), a sampling method that learns an adaptive policy to sequentially select k-space samples to optimize for target disease detection. On 6 out of 8 pathology classification tasks spanning the Knee, Brain, and Prostate MR scans, ASMR reaches within 2% of the performance of a fully sampled classifier while using only 8% of the k-space, as well as outperforming prior state-of-the-art work in k-space sampling such as EMRT, LOUPE, and DPS.
A Framework for Multi-modal Learning: Jointly Modeling Inter- & Intra-Modality Dependencies
May 27, 2024



Abstract:Supervised multi-modal learning involves mapping multiple modalities to a target label. Previous studies in this field have concentrated on capturing in isolation either the inter-modality dependencies (the relationships between different modalities and the label) or the intra-modality dependencies (the relationships within a single modality and the label). We argue that these conventional approaches that rely solely on either inter- or intra-modality dependencies may not be optimal in general. We view the multi-modal learning problem from the lens of generative models where we consider the target as a source of multiple modalities and the interaction between them. Towards that end, we propose inter- & intra-modality modeling (I2M2) framework, which captures and integrates both the inter- and intra-modality dependencies, leading to more accurate predictions. We evaluate our approach using real-world healthcare and vision-and-language datasets with state-of-the-art models, demonstrating superior performance over traditional methods focusing only on one type of modality dependency.
On Sensitivity and Robustness of Normalization Schemes to Input Distribution Shifts in Automatic MR Image Diagnosis
Jun 23, 2023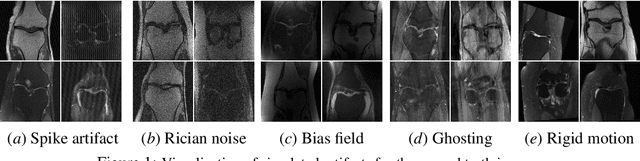
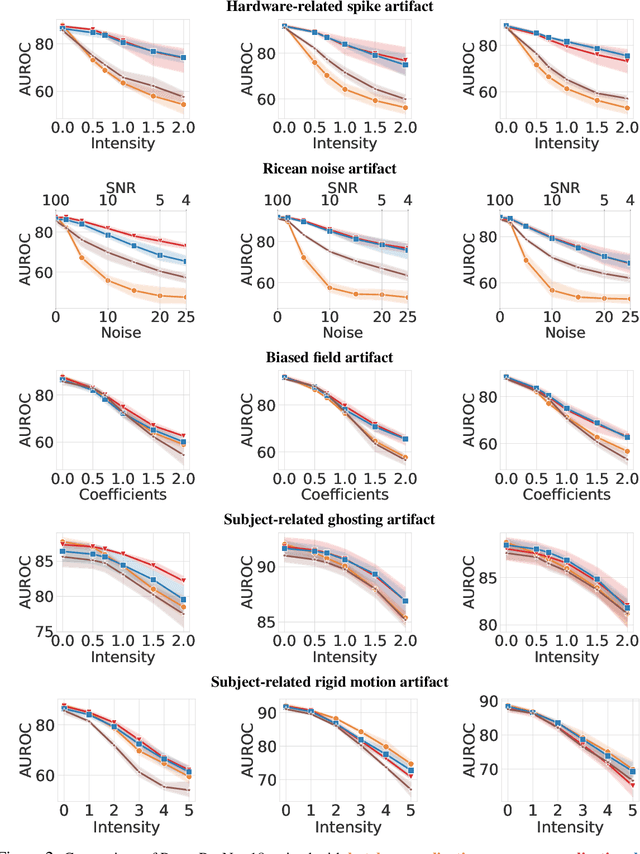
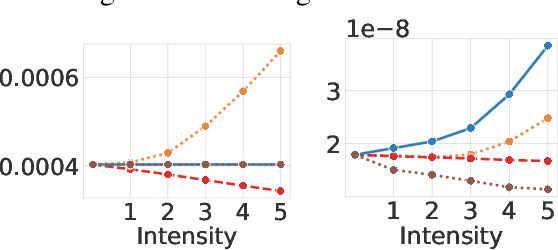
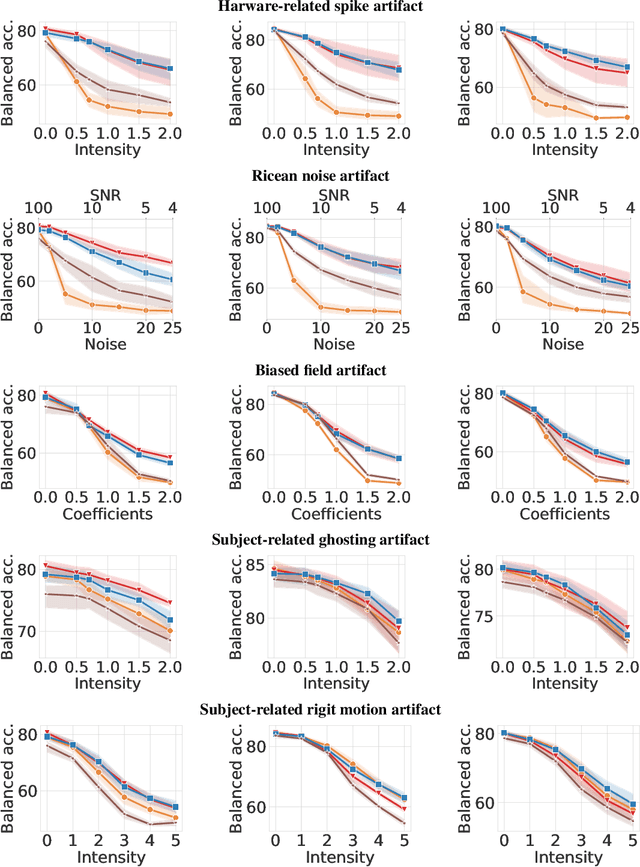
Abstract:Magnetic Resonance Imaging (MRI) is considered the gold standard of medical imaging because of the excellent soft-tissue contrast exhibited in the images reconstructed by the MRI pipeline, which in-turn enables the human radiologist to discern many pathologies easily. More recently, Deep Learning (DL) models have also achieved state-of-the-art performance in diagnosing multiple diseases using these reconstructed images as input. However, the image reconstruction process within the MRI pipeline, which requires the use of complex hardware and adjustment of a large number of scanner parameters, is highly susceptible to noise of various forms, resulting in arbitrary artifacts within the images. Furthermore, the noise distribution is not stationary and varies within a machine, across machines, and patients, leading to varying artifacts within the images. Unfortunately, DL models are quite sensitive to these varying artifacts as it leads to changes in the input data distribution between the training and testing phases. The lack of robustness of these models against varying artifacts impedes their use in medical applications where safety is critical. In this work, we focus on improving the generalization performance of these models in the presence of multiple varying artifacts that manifest due to the complexity of the MR data acquisition. In our experiments, we observe that Batch Normalization, a widely used technique during the training of DL models for medical image analysis, is a significant cause of performance degradation in these changing environments. As a solution, we propose to use other normalization techniques, such as Group Normalization and Layer Normalization (LN), to inject robustness into model performance against varying image artifacts. Through a systematic set of experiments, we show that GN and LN provide better accuracy for various MR artifacts and distribution shifts.
FastMRI Prostate: A Publicly Available, Biparametric MRI Dataset to Advance Machine Learning for Prostate Cancer Imaging
Apr 18, 2023Abstract:The fastMRI brain and knee dataset has enabled significant advances in exploring reconstruction methods for improving speed and image quality for Magnetic Resonance Imaging (MRI) via novel, clinically relevant reconstruction approaches. In this study, we describe the April 2023 expansion of the fastMRI dataset to include biparametric prostate MRI data acquired on a clinical population. The dataset consists of raw k-space and reconstructed images for T2-weighted and diffusion-weighted sequences along with slice-level labels that indicate the presence and grade of prostate cancer. As has been the case with fastMRI, increasing accessibility to raw prostate MRI data will further facilitate research in MR image reconstruction and evaluation with the larger goal of improving the utility of MRI for prostate cancer detection and evaluation. The dataset is available at https://fastmri.med.nyu.edu.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge