Daniel K Sodickson
DeepEMC-T2 Mapping: Deep Learning-Enabled T2 Mapping Based on Echo Modulation Curve Modeling
Feb 29, 2024Abstract:Purpose: Echo modulation curve (EMC) modeling can provide accurate and reproducible quantification of T2 relaxation times. The standard EMC-T2 mapping framework, however, requires sufficient echoes and cumbersome pixel-wise dictionary-matching steps. This work proposes a deep learning version of EMC-T2 mapping, called DeepEMC-T2 mapping, to efficiently estimate accurate T2 maps from fewer echoes without a dictionary. Methods: DeepEMC-T2 mapping was developed using a modified U-Net to estimate both T2 and Proton Density (PD) maps directly from multi-echo spin-echo (MESE) images. The modified U-Net employs several new features to improve the accuracy of T2/PD estimation. MESE datasets from 68 subjects were used for training and evaluation of the DeepEMC-T2 mapping technique. Multiple experiments were conducted to evaluate the impact of the proposed new features on DeepEMC-T2 mapping. Results: DeepEMC-T2 mapping achieved T2 estimation errors ranging from 3%-12% in different T2 ranges and 0.8%-1.7% for PD estimation with 10/7/5/3 echoes, which yielded more accurate parameter estimation than standard EMC-T2 mapping. The new features proposed in DeepEMC-T2 mapping enabled improved parameter estimation. The use of a larger echo spacing with fewer echoes can maintain the accuracy of T2 and PD estimations while reducing the number of 180-degree refocusing pulses. Conclusions: DeepEMC-T2 mapping enables simplified, efficient, and accurate T2 quantification directly from MESE images without a time-consuming dictionary-matching step and requires fewer echoes. This allows for increased volumetric coverage and/or decreased SAR by reducing the number of 180-degree refocusing pulses.
FastMRI Prostate: A Publicly Available, Biparametric MRI Dataset to Advance Machine Learning for Prostate Cancer Imaging
Apr 18, 2023Abstract:The fastMRI brain and knee dataset has enabled significant advances in exploring reconstruction methods for improving speed and image quality for Magnetic Resonance Imaging (MRI) via novel, clinically relevant reconstruction approaches. In this study, we describe the April 2023 expansion of the fastMRI dataset to include biparametric prostate MRI data acquired on a clinical population. The dataset consists of raw k-space and reconstructed images for T2-weighted and diffusion-weighted sequences along with slice-level labels that indicate the presence and grade of prostate cancer. As has been the case with fastMRI, increasing accessibility to raw prostate MRI data will further facilitate research in MR image reconstruction and evaluation with the larger goal of improving the utility of MRI for prostate cancer detection and evaluation. The dataset is available at https://fastmri.med.nyu.edu.
Learning a Variational Network for Reconstruction of Accelerated MRI Data
Apr 03, 2017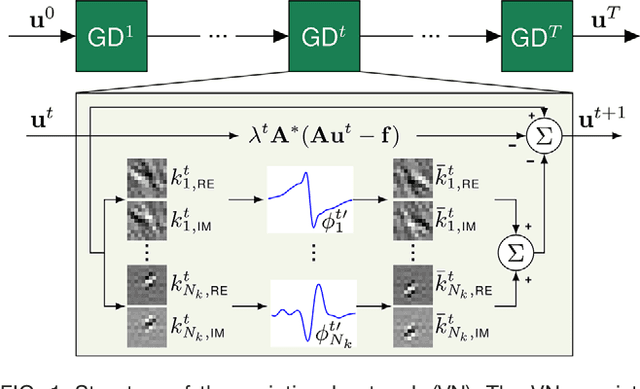
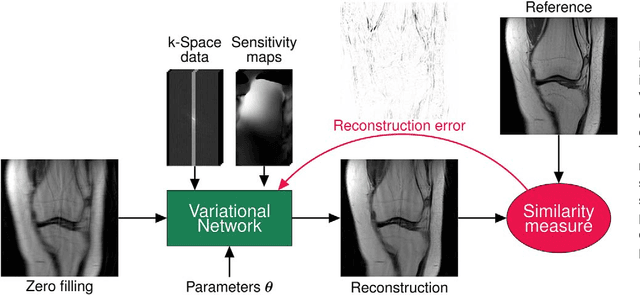
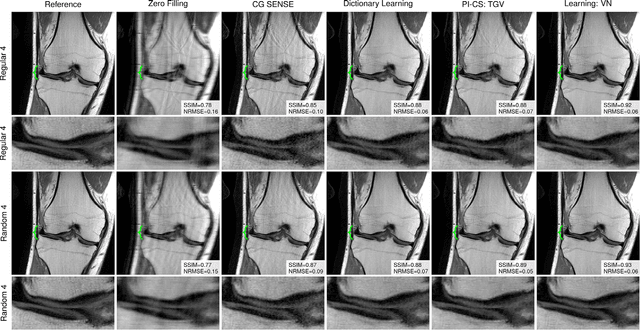
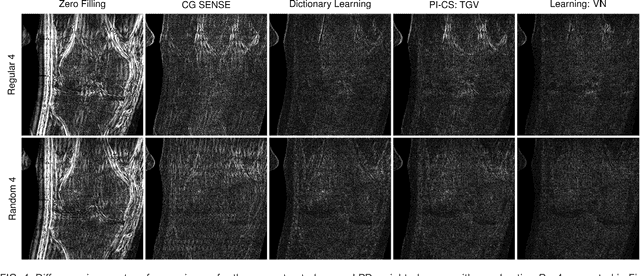
Abstract:Purpose: To allow fast and high-quality reconstruction of clinical accelerated multi-coil MR data by learning a variational network that combines the mathematical structure of variational models with deep learning. Theory and Methods: Generalized compressed sensing reconstruction formulated as a variational model is embedded in an unrolled gradient descent scheme. All parameters of this formulation, including the prior model defined by filter kernels and activation functions as well as the data term weights, are learned during an offline training procedure. The learned model can then be applied online to previously unseen data. Results: The variational network approach is evaluated on a clinical knee imaging protocol. The variational network reconstructions outperform standard reconstruction algorithms in terms of image quality and residual artifacts for all tested acceleration factors and sampling patterns. Conclusion: Variational network reconstructions preserve the natural appearance of MR images as well as pathologies that were not included in the training data set. Due to its high computational performance, i.e., reconstruction time of 193 ms on a single graphics card, and the omission of parameter tuning once the network is trained, this new approach to image reconstruction can easily be integrated into clinical workflow.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge