Luoyao Chen
Kolmogorov-Arnold Representation for Symplectic Learning: Advancing Hamiltonian Neural Networks
Aug 26, 2025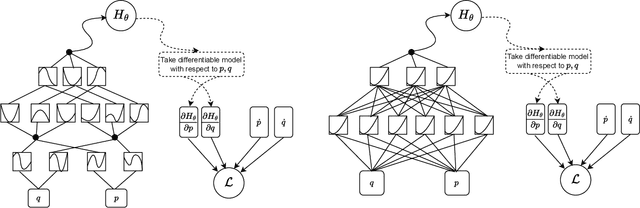
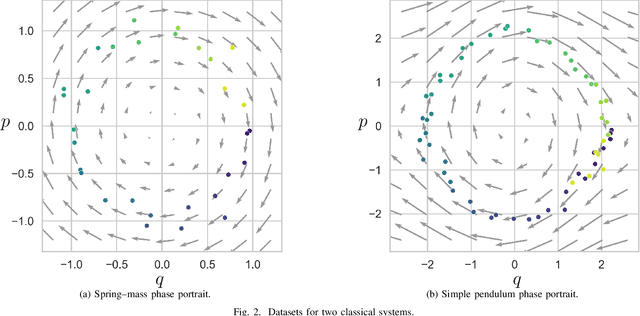
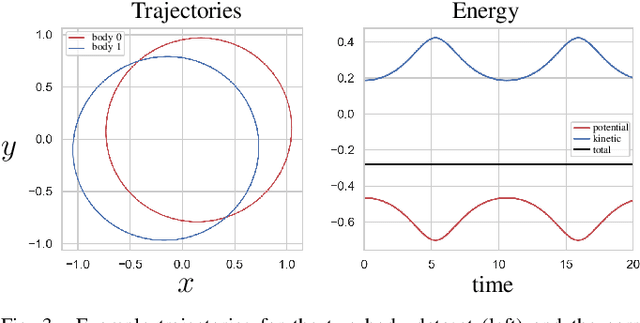
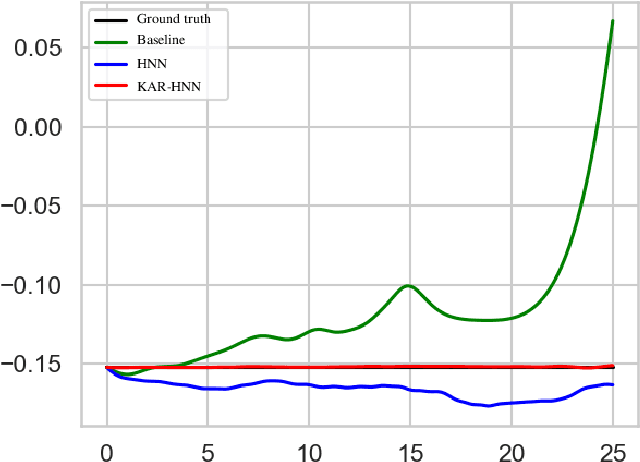
Abstract:We propose a Kolmogorov-Arnold Representation-based Hamiltonian Neural Network (KAR-HNN) that replaces the Multilayer Perceptrons (MLPs) with univariate transformations. While Hamiltonian Neural Networks (HNNs) ensure energy conservation by learning Hamiltonian functions directly from data, existing implementations, often relying on MLPs, cause hypersensitivity to the hyperparameters while exploring complex energy landscapes. Our approach exploits the localized function approximations to better capture high-frequency and multi-scale dynamics, reducing energy drift and improving long-term predictive stability. The networks preserve the symplectic form of Hamiltonian systems, and thus maintain interpretability and physical consistency. After assessing KAR-HNN on four benchmark problems including spring-mass, simple pendulum, two- and three-body problem, we foresee its effectiveness for accurate and stable modeling of realistic physical processes often at high dimensions and with few known parameters.
Fine-Tuning In-House Large Language Models to Infer Differential Diagnosis from Radiology Reports
Oct 11, 2024



Abstract:Radiology reports summarize key findings and differential diagnoses derived from medical imaging examinations. The extraction of differential diagnoses is crucial for downstream tasks, including patient management and treatment planning. However, the unstructured nature of these reports, characterized by diverse linguistic styles and inconsistent formatting, presents significant challenges. Although proprietary large language models (LLMs) such as GPT-4 can effectively retrieve clinical information, their use is limited in practice by high costs and concerns over the privacy of protected health information (PHI). This study introduces a pipeline for developing in-house LLMs tailored to identify differential diagnoses from radiology reports. We first utilize GPT-4 to create 31,056 labeled reports, then fine-tune open source LLM using this dataset. Evaluated on a set of 1,067 reports annotated by clinicians, the proposed model achieves an average F1 score of 92.1\%, which is on par with GPT-4 (90.8\%). Through this study, we provide a methodology for constructing in-house LLMs that: match the performance of GPT, reduce dependence on expensive proprietary models, and enhance the privacy and security of PHI.
Shifting to Machine Supervision: Annotation-Efficient Semi and Self-Supervised Learning for Automatic Medical Image Segmentation and Classification
Nov 17, 2023Abstract:Advancements in clinical treatment and research are limited by supervised learning techniques that rely on large amounts of annotated data, an expensive task requiring many hours of clinical specialists' time. In this paper, we propose using self-supervised and semi-supervised learning. These techniques perform an auxiliary task that is label-free, scaling up machine-supervision is easier compared with fully-supervised techniques. This paper proposes S4MI (Self-Supervision and Semi-Supervision for Medical Imaging), our pipeline to leverage advances in self and semi-supervision learning. We benchmark them on three medical imaging datasets to analyze their efficacy for classification and segmentation. This advancement in self-supervised learning with 10% annotation performed better than 100% annotation for the classification of most datasets. The semi-supervised approach yielded favorable outcomes for segmentation, outperforming the fully-supervised approach by using 50% fewer labels in all three datasets.
Enhancing Medical Image Segmentation: Optimizing Cross-Entropy Weights and Post-Processing with Autoencoders
Aug 21, 2023



Abstract:The task of medical image segmentation presents unique challenges, necessitating both localized and holistic semantic understanding to accurately delineate areas of interest, such as critical tissues or aberrant features. This complexity is heightened in medical image segmentation due to the high degree of inter-class similarities, intra-class variations, and possible image obfuscation. The segmentation task further diversifies when considering the study of histopathology slides for autoimmune diseases like dermatomyositis. The analysis of cell inflammation and interaction in these cases has been less studied due to constraints in data acquisition pipelines. Despite the progressive strides in medical science, we lack a comprehensive collection of autoimmune diseases. As autoimmune diseases globally escalate in prevalence and exhibit associations with COVID-19, their study becomes increasingly essential. While there is existing research that integrates artificial intelligence in the analysis of various autoimmune diseases, the exploration of dermatomyositis remains relatively underrepresented. In this paper, we present a deep-learning approach tailored for Medical image segmentation. Our proposed method outperforms the current state-of-the-art techniques by an average of 12.26% for U-Net and 12.04% for U-Net++ across the ResNet family of encoders on the dermatomyositis dataset. Furthermore, we probe the importance of optimizing loss function weights and benchmark our methodology on three challenging medical image segmentation tasks
Egocentric Prediction of Action Target in 3D
Mar 24, 2022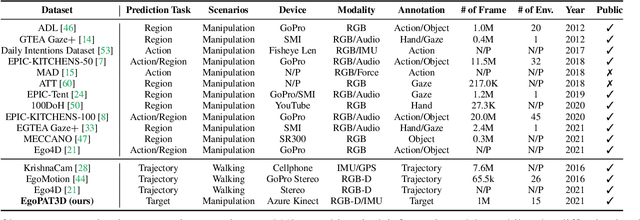
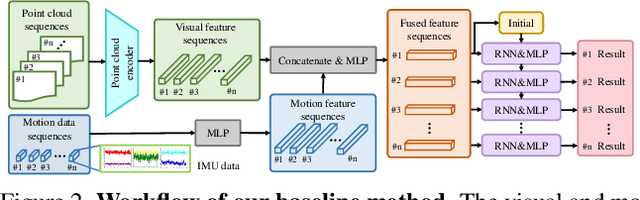
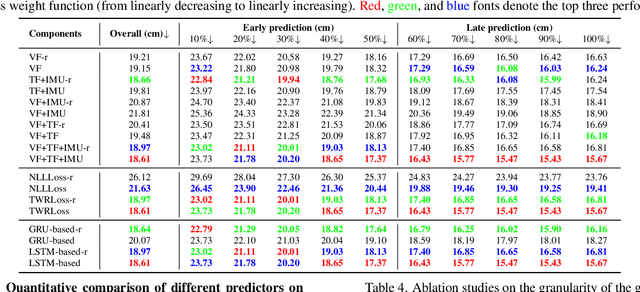
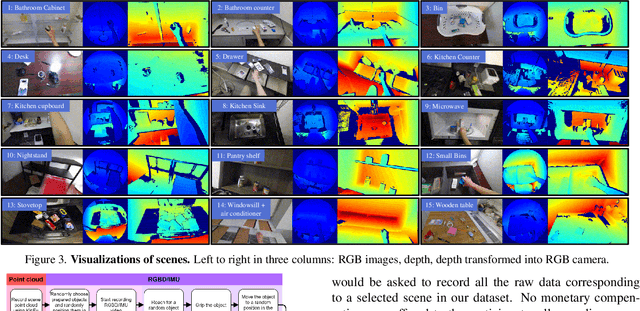
Abstract:We are interested in anticipating as early as possible the target location of a person's object manipulation action in a 3D workspace from egocentric vision. It is important in fields like human-robot collaboration, but has not yet received enough attention from vision and learning communities. To stimulate more research on this challenging egocentric vision task, we propose a large multimodality dataset of more than 1 million frames of RGB-D and IMU streams, and provide evaluation metrics based on our high-quality 2D and 3D labels from semi-automatic annotation. Meanwhile, we design baseline methods using recurrent neural networks and conduct various ablation studies to validate their effectiveness. Our results demonstrate that this new task is worthy of further study by researchers in robotics, vision, and learning communities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge