Jinqian Pan
DTC: A Deformable Transposed Convolution Module for Medical Image Segmentation
Jan 25, 2026Abstract:In medical image segmentation, particularly in UNet-like architectures, upsampling is primarily used to transform smaller feature maps into larger ones, enabling feature fusion between encoder and decoder features and supporting multi-scale prediction. Conventional upsampling methods, such as transposed convolution and linear interpolation, operate on fixed positions: transposed convolution applies kernel elements to predetermined pixel or voxel locations, while linear interpolation assigns values based on fixed coordinates in the original feature map. These fixed-position approaches may fail to capture structural information beyond predefined sampling positions and can lead to artifacts or loss of detail. Inspired by deformable convolutions, we propose a novel upsampling method, Deformable Transposed Convolution (DTC), which learns dynamic coordinates (i.e., sampling positions) to generate high-resolution feature maps for both 2D and 3D medical image segmentation tasks. Experiments on 3D (e.g., BTCV15) and 2D datasets (e.g., ISIC18, BUSI) demonstrate that DTC can be effectively integrated into existing medical image segmentation models, consistently improving the decoder's feature reconstruction and detail recovery capability.
Natural Language Generation in Healthcare: A Review of Methods and Applications
May 07, 2025Abstract:Natural language generation (NLG) is the key technology to achieve generative artificial intelligence (AI). With the breakthroughs in large language models (LLMs), NLG has been widely used in various medical applications, demonstrating the potential to enhance clinical workflows, support clinical decision-making, and improve clinical documentation. Heterogeneous and diverse medical data modalities, such as medical text, images, and knowledge bases, are utilized in NLG. Researchers have proposed many generative models and applied them in a number of healthcare applications. There is a need for a comprehensive review of NLG methods and applications in the medical domain. In this study, we systematically reviewed 113 scientific publications from a total of 3,988 NLG-related articles identified using a literature search, focusing on data modality, model architecture, clinical applications, and evaluation methods. Following PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines, we categorize key methods, identify clinical applications, and assess their capabilities, limitations, and emerging challenges. This timely review covers the key NLG technologies and medical applications and provides valuable insights for future studies to leverage NLG to transform medical discovery and healthcare.
Shifting to Machine Supervision: Annotation-Efficient Semi and Self-Supervised Learning for Automatic Medical Image Segmentation and Classification
Nov 17, 2023Abstract:Advancements in clinical treatment and research are limited by supervised learning techniques that rely on large amounts of annotated data, an expensive task requiring many hours of clinical specialists' time. In this paper, we propose using self-supervised and semi-supervised learning. These techniques perform an auxiliary task that is label-free, scaling up machine-supervision is easier compared with fully-supervised techniques. This paper proposes S4MI (Self-Supervision and Semi-Supervision for Medical Imaging), our pipeline to leverage advances in self and semi-supervision learning. We benchmark them on three medical imaging datasets to analyze their efficacy for classification and segmentation. This advancement in self-supervised learning with 10% annotation performed better than 100% annotation for the classification of most datasets. The semi-supervised approach yielded favorable outcomes for segmentation, outperforming the fully-supervised approach by using 50% fewer labels in all three datasets.
Enhancing Medical Image Segmentation: Optimizing Cross-Entropy Weights and Post-Processing with Autoencoders
Aug 21, 2023



Abstract:The task of medical image segmentation presents unique challenges, necessitating both localized and holistic semantic understanding to accurately delineate areas of interest, such as critical tissues or aberrant features. This complexity is heightened in medical image segmentation due to the high degree of inter-class similarities, intra-class variations, and possible image obfuscation. The segmentation task further diversifies when considering the study of histopathology slides for autoimmune diseases like dermatomyositis. The analysis of cell inflammation and interaction in these cases has been less studied due to constraints in data acquisition pipelines. Despite the progressive strides in medical science, we lack a comprehensive collection of autoimmune diseases. As autoimmune diseases globally escalate in prevalence and exhibit associations with COVID-19, their study becomes increasingly essential. While there is existing research that integrates artificial intelligence in the analysis of various autoimmune diseases, the exploration of dermatomyositis remains relatively underrepresented. In this paper, we present a deep-learning approach tailored for Medical image segmentation. Our proposed method outperforms the current state-of-the-art techniques by an average of 12.26% for U-Net and 12.04% for U-Net++ across the ResNet family of encoders on the dermatomyositis dataset. Furthermore, we probe the importance of optimizing loss function weights and benchmark our methodology on three challenging medical image segmentation tasks
Low-Light Image and Video Enhancement: A Comprehensive Survey and Beyond
Dec 21, 2022Abstract:This paper presents a comprehensive survey of low-light image and video enhancement. We begin with the challenging mixed over-/under-exposed images, which are under-performed by existing methods. To this end, we propose two variants of the SICE dataset named SICE_Grad and SICE_Mix. Next, we introduce Night Wenzhou, a large-scale, high-resolution video dataset, to address the issue of the lack of a low-light video dataset that discount the use of low-light image enhancement (LLIE) to videos. The Night Wenzhou dataset is challenging since it consists of fast-moving aerial scenes and streetscapes with varying illuminations and degradation. We conduct extensive key technique analysis and experimental comparisons for representative LLIE approaches using these newly proposed datasets and the current benchmark datasets. Finally, we address unresolved issues and propose future research topics for the LLIE community.
PointNorm: Normalization is All You Need for Point Cloud Analysis
Jul 13, 2022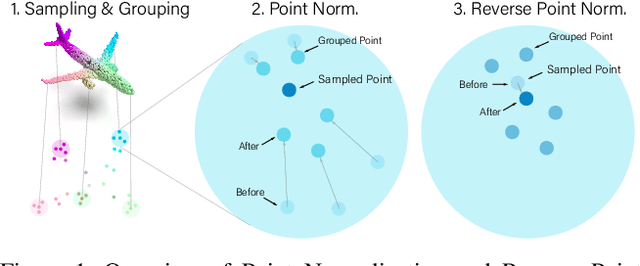
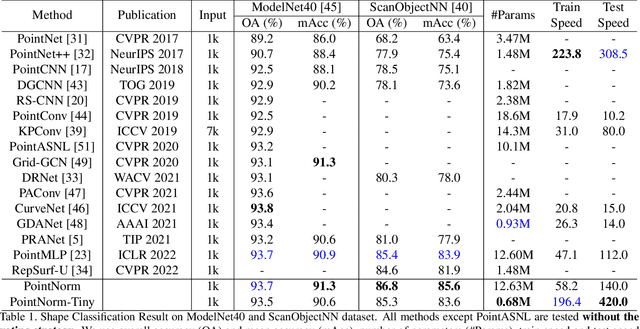
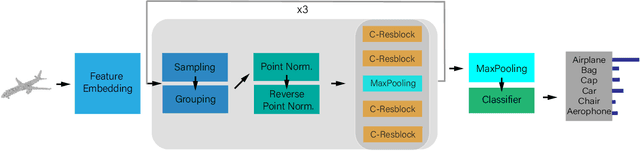
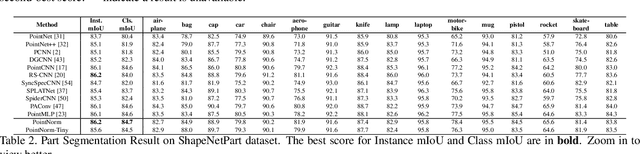
Abstract:Point cloud analysis is challenging due to the irregularity of the point cloud data structure. Existing works typically employ the ad-hoc sampling-grouping operation of PointNet++, followed by sophisticated local and/or global feature extractors for leveraging the 3D geometry of the point cloud. Unfortunately, those intricate hand-crafted model designs have led to poor inference latency and performance saturation in the last few years. In this paper, we point out that the classical sampling-grouping operations on the irregular point cloud cause learning difficulty for the subsequent MLP layers. To reduce the irregularity of the point cloud, we introduce a DualNorm module after the sampling-grouping operation. The DualNorm module consists of Point Normalization, which normalizes the grouped points to the sampled points, and Reverse Point Normalization, which normalizes the sampled points to the grouped points. The proposed PointNorm utilizes local mean and global standard deviation to benefit from both local and global features while maintaining a faithful inference speed. Experiments on point cloud classification show that we achieved state-of-the-art accuracy on ModelNet40 and ScanObjectNN datasets. We also generalize our model to point cloud part segmentation and demonstrate competitive performance on the ShapeNetPart dataset. Code is available at https://github.com/ShenZheng2000/PointNorm-for-Point-Cloud-Analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge