Jacopo Cirrone
Exploring Intrinsic Properties of Medical Images for Self-Supervised Binary Semantic Segmentation
Feb 04, 2024Abstract:Recent advancements in self-supervised learning have unlocked the potential to harness unlabeled data for auxiliary tasks, facilitating the learning of beneficial priors. This has been particularly advantageous in fields like medical image analysis, where labeled data are scarce. Although effective for classification tasks, this methodology has shown limitations in more complex applications, such as medical image segmentation. In this paper, we introduce Medical imaging Enhanced with Dynamic Self-Adaptive Semantic Segmentation (MedSASS), a dedicated self-supervised framework tailored for medical image segmentation. We evaluate MedSASS against existing state-of-the-art methods across four diverse medical datasets, showcasing its superiority. MedSASS outperforms existing CNN-based self-supervised methods by 3.83% and matches the performance of ViT-based methods. Furthermore, when MedSASS is trained end-to-end, covering both encoder and decoder, it demonstrates significant improvements of 14.4% for CNNs and 6% for ViT-based architectures compared to existing state-of-the-art self-supervised strategies.
Free Form Medical Visual Question Answering in Radiology
Jan 23, 2024Abstract:Visual Question Answering (VQA) in the medical domain presents a unique, interdisciplinary challenge, combining fields such as Computer Vision, Natural Language Processing, and Knowledge Representation. Despite its importance, research in medical VQA has been scant, only gaining momentum since 2018. Addressing this gap, our research delves into the effective representation of radiology images and the joint learning of multimodal representations, surpassing existing methods. We innovatively augment the SLAKE dataset, enabling our model to respond to a more diverse array of questions, not limited to the immediate content of radiology or pathology images. Our model achieves a top-1 accuracy of 79.55\% with a less complex architecture, demonstrating comparable performance to current state-of-the-art models. This research not only advances medical VQA but also opens avenues for practical applications in diagnostic settings.
Shifting to Machine Supervision: Annotation-Efficient Semi and Self-Supervised Learning for Automatic Medical Image Segmentation and Classification
Nov 17, 2023Abstract:Advancements in clinical treatment and research are limited by supervised learning techniques that rely on large amounts of annotated data, an expensive task requiring many hours of clinical specialists' time. In this paper, we propose using self-supervised and semi-supervised learning. These techniques perform an auxiliary task that is label-free, scaling up machine-supervision is easier compared with fully-supervised techniques. This paper proposes S4MI (Self-Supervision and Semi-Supervision for Medical Imaging), our pipeline to leverage advances in self and semi-supervision learning. We benchmark them on three medical imaging datasets to analyze their efficacy for classification and segmentation. This advancement in self-supervised learning with 10% annotation performed better than 100% annotation for the classification of most datasets. The semi-supervised approach yielded favorable outcomes for segmentation, outperforming the fully-supervised approach by using 50% fewer labels in all three datasets.
Enhancing Medical Image Segmentation: Optimizing Cross-Entropy Weights and Post-Processing with Autoencoders
Aug 21, 2023



Abstract:The task of medical image segmentation presents unique challenges, necessitating both localized and holistic semantic understanding to accurately delineate areas of interest, such as critical tissues or aberrant features. This complexity is heightened in medical image segmentation due to the high degree of inter-class similarities, intra-class variations, and possible image obfuscation. The segmentation task further diversifies when considering the study of histopathology slides for autoimmune diseases like dermatomyositis. The analysis of cell inflammation and interaction in these cases has been less studied due to constraints in data acquisition pipelines. Despite the progressive strides in medical science, we lack a comprehensive collection of autoimmune diseases. As autoimmune diseases globally escalate in prevalence and exhibit associations with COVID-19, their study becomes increasingly essential. While there is existing research that integrates artificial intelligence in the analysis of various autoimmune diseases, the exploration of dermatomyositis remains relatively underrepresented. In this paper, we present a deep-learning approach tailored for Medical image segmentation. Our proposed method outperforms the current state-of-the-art techniques by an average of 12.26% for U-Net and 12.04% for U-Net++ across the ResNet family of encoders on the dermatomyositis dataset. Furthermore, we probe the importance of optimizing loss function weights and benchmark our methodology on three challenging medical image segmentation tasks
Efficient Representation Learning for Healthcare with Cross-Architectural Self-Supervision
Aug 19, 2023Abstract:In healthcare and biomedical applications, extreme computational requirements pose a significant barrier to adopting representation learning. Representation learning can enhance the performance of deep learning architectures by learning useful priors from limited medical data. However, state-of-the-art self-supervised techniques suffer from reduced performance when using smaller batch sizes or shorter pretraining epochs, which are more practical in clinical settings. We present Cross Architectural - Self Supervision (CASS) in response to this challenge. This novel siamese self-supervised learning approach synergistically leverages Transformer and Convolutional Neural Networks (CNN) for efficient learning. Our empirical evaluation demonstrates that CASS-trained CNNs and Transformers outperform existing self-supervised learning methods across four diverse healthcare datasets. With only 1% labeled data for finetuning, CASS achieves a 3.8% average improvement; with 10% labeled data, it gains 5.9%; and with 100% labeled data, it reaches a remarkable 10.13% enhancement. Notably, CASS reduces pretraining time by 69% compared to state-of-the-art methods, making it more amenable to clinical implementation. We also demonstrate that CASS is considerably more robust to variations in batch size and pretraining epochs, making it a suitable candidate for machine learning in healthcare applications.
Cross-Architectural Positive Pairs improve the effectiveness of Self-Supervised Learning
Jan 27, 2023

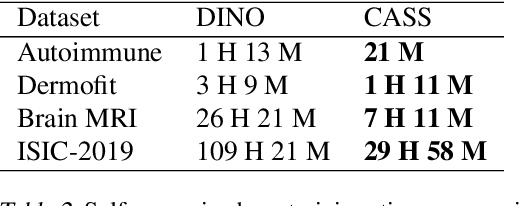

Abstract:Existing self-supervised techniques have extreme computational requirements and suffer a substantial drop in performance with a reduction in batch size or pretraining epochs. This paper presents Cross Architectural - Self Supervision (CASS), a novel self-supervised learning approach that leverages Transformer and CNN simultaneously. Compared to the existing state-of-the-art self-supervised learning approaches, we empirically show that CASS-trained CNNs and Transformers across four diverse datasets gained an average of 3.8% with 1% labeled data, 5.9% with 10% labeled data, and 10.13% with 100% labeled data while taking 69% less time. We also show that CASS is much more robust to changes in batch size and training epochs than existing state-of-the-art self-supervised learning approaches. We have open-sourced our code at https://github.com/pranavsinghps1/CASS.
Prediction of drug effectiveness in rheumatoid arthritis patients based on machine learning algorithms
Oct 18, 2022



Abstract:Rheumatoid arthritis (RA) is an autoimmune condition caused when patients' immune system mistakenly targets their own tissue. Machine learning (ML) has the potential to identify patterns in patient electronic health records (EHR) to forecast the best clinical treatment to improve patient outcomes. This study introduced a Drug Response Prediction (DRP) framework with two main goals: 1) design a data processing pipeline to extract information from tabular clinical data, and then preprocess it for functional use, and 2) predict RA patient's responses to drugs and evaluate classification models' performance. We propose a novel two-stage ML framework based on European Alliance of Associations for Rheumatology (EULAR) criteria cutoffs to model drug effectiveness. Our model Stacked-Ensemble DRP was developed and cross-validated using data from 425 RA patients. The evaluation used a subset of 124 patients (30%) from the same data source. In the evaluation of the test set, two-stage DRP leads to improved classification accuracy over other end-to-end classification models for binary classification. Our proposed method provides a complete pipeline to predict disease activity scores and identify the group that does not respond well to anti-TNF treatments, thus showing promise in supporting clinical decisions based on EHR information.
A Data-Efficient Deep Learning Framework for Segmentation and Classification of Histopathology Images
Jul 16, 2022
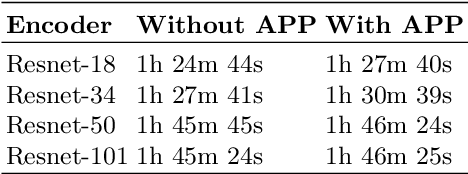

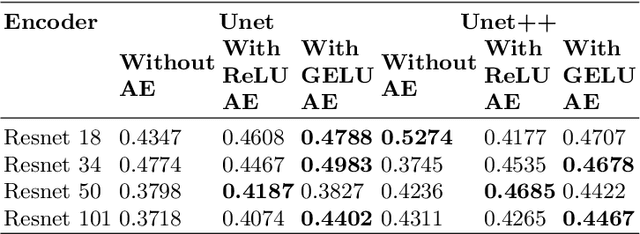
Abstract:The current study of cell architecture of inflammation in histopathology images commonly performed for diagnosis and research purposes excludes a lot of information available on the biopsy slide. In autoimmune diseases, major outstanding research questions remain regarding which cell types participate in inflammation at the tissue level,and how they interact with each other. While these questions can be partially answered using traditional methods, artificial intelligence approaches for segmentation and classification provide a much more efficient method to understand the architecture of inflammation in autoimmune disease, holding a great promise for novel insights. In this paper, we empirically develop deep learning approaches that uses dermatomyositis biopsies of human tissue to detect and identify inflammatory cells. Our approach improves classification performance by 26% and segmentation performance by 5%. We also propose a novel post-processing autoencoder architecture that improves segmentation performance by an additional 3%. We have open-sourced our approach and architecture at https://github.com/pranavsinghps1/DEDL
CASS: Cross Architectural Self-Supervision for Medical Image Analysis
Jun 23, 2022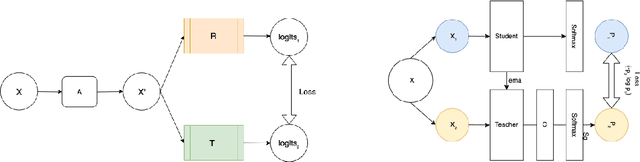
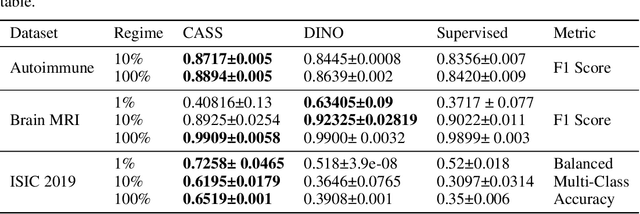

Abstract:Recent advances in Deep Learning and Computer Vision have alleviated many of the bottlenecks, allowing algorithms to be label-free with better performance. Specifically, Transformers provide a global perspective of the image, which Convolutional Neural Networks (CNN) lack by design. Here we present Cross Architectural Self-Supervision, a novel self-supervised learning approach which leverages transformers and CNN simultaneously, while also being computationally accessible to general practitioners via easily available cloud services. Compared to existing state-of-the-art self-supervised learning approaches, we empirically show CASS trained CNNs, and Transformers gained an average of 8.5% with 100% labelled data, 7.3% with 10% labelled data, and 11.5% with 1% labelled data, across three diverse datasets. Notably, one of the employed datasets included histopathology slides of an autoimmune disease, a topic underrepresented in Medical Imaging and has minimal data. In addition, our findings reveal that CASS is twice as efficient as other state-of-the-art methods in terms of training time. The code is open source and is available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge