Shravan Chaudhari
Shared LoRA Subspaces for almost Strict Continual Learning
Feb 05, 2026Abstract:Adapting large pretrained models to new tasks efficiently and continually is crucial for real-world deployment but remains challenging due to catastrophic forgetting and the high cost of retraining. While parameter-efficient tuning methods like low rank adaptation (LoRA) reduce computational demands, they lack mechanisms for strict continual learning and knowledge integration, without relying on data replay, or multiple adapters. We propose Share, a novel approach to parameter efficient continual finetuning that learns and dynamically updates a single, shared low-rank subspace, enabling seamless adaptation across multiple tasks and modalities. Share constructs a foundational subspace that extracts core knowledge from past tasks and incrementally integrates new information by identifying essential subspace directions. Knowledge from each new task is incorporated into this evolving subspace, facilitating forward knowledge transfer, while minimizing catastrophic interference. This approach achieves up to 100x parameter reduction and 281x memory savings over traditional LoRA methods, maintaining performance comparable to jointly trained models. A single Share model can replace hundreds of task-specific LoRA adapters, supporting scalable, asynchronous continual learning. Experiments across image classification, natural language understanding, 3D pose estimation, and text-to-image generation validate its effectiveness, making Share a practical and scalable solution for lifelong learning in large-scale AI systems.
SpIDER: Spatially Informed Dense Embedding Retrieval for Software Issue Localization
Dec 18, 2025Abstract:Retrieving code units (e.g., files, classes, functions) that are semantically relevant to a given user query, bug report, or feature request from large codebases is a fundamental challenge for LLM-based coding agents. Agentic approaches typically employ sparse retrieval methods like BM25 or dense embedding strategies to identify relevant units. While embedding-based approaches can outperform BM25 by large margins, they often lack exploration of the codebase and underutilize its underlying graph structure. To address this, we propose SpIDER (Spatially Informed Dense Embedding Retrieval), an enhanced dense retrieval approach that incorporates LLM-based reasoning over auxiliary context obtained through graph-based exploration of the codebase. Empirical results show that SpIDER consistently improves dense retrieval performance across several programming languages.
Multimodal LLM Augmented Reasoning for Interpretable Visual Perception Analysis
Apr 16, 2025Abstract:In this paper, we advance the study of AI-augmented reasoning in the context of Human-Computer Interaction (HCI), psychology and cognitive science, focusing on the critical task of visual perception. Specifically, we investigate the applicability of Multimodal Large Language Models (MLLMs) in this domain. To this end, we leverage established principles and explanations from psychology and cognitive science related to complexity in human visual perception. We use them as guiding principles for the MLLMs to compare and interprete visual content. Our study aims to benchmark MLLMs across various explainability principles relevant to visual perception. Unlike recent approaches that primarily employ advanced deep learning models to predict complexity metrics from visual content, our work does not seek to develop a mere new predictive model. Instead, we propose a novel annotation-free analytical framework to assess utility of MLLMs as cognitive assistants for HCI tasks, using visual perception as a case study. The primary goal is to pave the way for principled study in quantifying and evaluating the interpretability of MLLMs for applications in improving human reasoning capability and uncovering biases in existing perception datasets annotated by humans.
Between Linear and Sinusoidal: Rethinking the Time Encoder in Dynamic Graph Learning
Apr 10, 2025
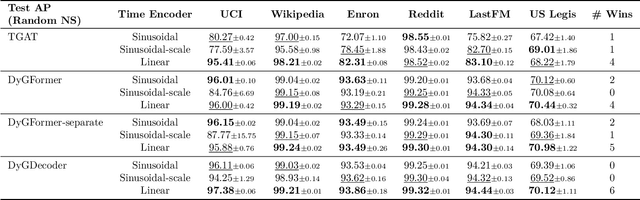
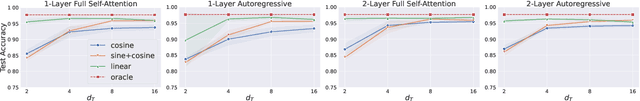

Abstract:Dynamic graph learning is essential for applications involving temporal networks and requires effective modeling of temporal relationships. Seminal attention-based models like TGAT and DyGFormer rely on sinusoidal time encoders to capture temporal relationships between edge events. In this paper, we study a simpler alternative: the linear time encoder, which avoids temporal information loss caused by sinusoidal functions and reduces the need for high dimensional time encoders. We show that the self-attention mechanism can effectively learn to compute time spans from linear time encodings and extract relevant temporal patterns. Through extensive experiments on six dynamic graph datasets, we demonstrate that the linear time encoder improves the performance of TGAT and DyGFormer in most cases. Moreover, the linear time encoder can lead to significant savings in model parameters with minimal performance loss. For example, compared to a 100-dimensional sinusoidal time encoder, TGAT with a 2-dimensional linear time encoder saves 43% of parameters and achieves higher average precision on five datasets. These results can be readily used to positively impact the design choices of a wide variety of dynamic graph learning architectures. The experimental code is available at: https://github.com/hsinghuan/dg-linear-time.git.
EigenLoRAx: Recycling Adapters to Find Principal Subspaces for Resource-Efficient Adaptation and Inference
Feb 07, 2025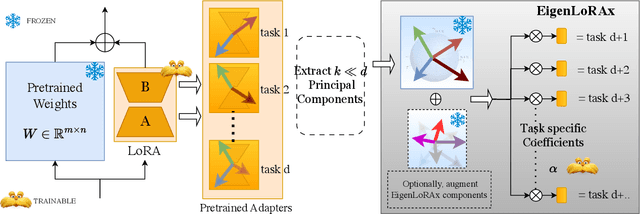
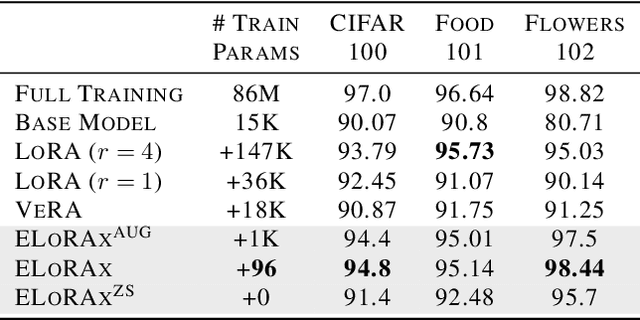
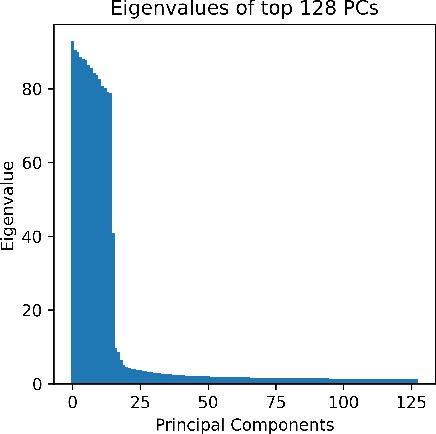
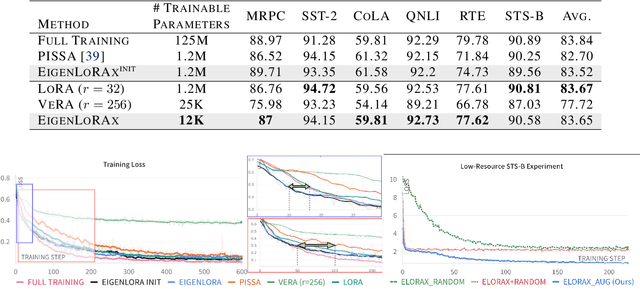
Abstract:The rapid growth of large models has raised concerns about their environmental impact and equity in accessibility due to significant computational costs. Low-Rank Adapters (LoRA) offer a lightweight solution for finetuning large models, resulting in an abundance of publicly available adapters tailored to diverse domains. We ask: Can these pretrained adapters be leveraged to further streamline adaptation to new tasks while addressing these challenges? We introduce EigenLoRAx, a parameter-efficient finetuning method that recycles existing adapters to create a principal subspace aligned with their shared domain knowledge which can be further augmented with orthogonal basis vectors in low-resource scenarios. This enables rapid adaptation to new tasks by learning only lightweight coefficients on the principal components of the subspace - eliminating the need to finetune entire adapters. EigenLoRAx requires significantly fewer parameters and memory, improving efficiency for both training and inference. Our method demonstrates strong performance across diverse domains and tasks, offering a scalable for edge-based applications, personalization, and equitable deployment of large models in resource-constrained environments.
Novel Node Category Detection Under Subpopulation Shift
Apr 01, 2024



Abstract:In real-world graph data, distribution shifts can manifest in various ways, such as the emergence of new categories and changes in the relative proportions of existing categories. It is often important to detect nodes of novel categories under such distribution shifts for safety or insight discovery purposes. We introduce a new approach, Recall-Constrained Optimization with Selective Link Prediction (RECO-SLIP), to detect nodes belonging to novel categories in attributed graphs under subpopulation shifts. By integrating a recall-constrained learning framework with a sample-efficient link prediction mechanism, RECO-SLIP addresses the dual challenges of resilience against subpopulation shifts and the effective exploitation of graph structure. Our extensive empirical evaluation across multiple graph datasets demonstrates the superior performance of RECO-SLIP over existing methods.
Shifting to Machine Supervision: Annotation-Efficient Semi and Self-Supervised Learning for Automatic Medical Image Segmentation and Classification
Nov 17, 2023Abstract:Advancements in clinical treatment and research are limited by supervised learning techniques that rely on large amounts of annotated data, an expensive task requiring many hours of clinical specialists' time. In this paper, we propose using self-supervised and semi-supervised learning. These techniques perform an auxiliary task that is label-free, scaling up machine-supervision is easier compared with fully-supervised techniques. This paper proposes S4MI (Self-Supervision and Semi-Supervision for Medical Imaging), our pipeline to leverage advances in self and semi-supervision learning. We benchmark them on three medical imaging datasets to analyze their efficacy for classification and segmentation. This advancement in self-supervised learning with 10% annotation performed better than 100% annotation for the classification of most datasets. The semi-supervised approach yielded favorable outcomes for segmentation, outperforming the fully-supervised approach by using 50% fewer labels in all three datasets.
Enhancing Medical Image Segmentation: Optimizing Cross-Entropy Weights and Post-Processing with Autoencoders
Aug 21, 2023



Abstract:The task of medical image segmentation presents unique challenges, necessitating both localized and holistic semantic understanding to accurately delineate areas of interest, such as critical tissues or aberrant features. This complexity is heightened in medical image segmentation due to the high degree of inter-class similarities, intra-class variations, and possible image obfuscation. The segmentation task further diversifies when considering the study of histopathology slides for autoimmune diseases like dermatomyositis. The analysis of cell inflammation and interaction in these cases has been less studied due to constraints in data acquisition pipelines. Despite the progressive strides in medical science, we lack a comprehensive collection of autoimmune diseases. As autoimmune diseases globally escalate in prevalence and exhibit associations with COVID-19, their study becomes increasingly essential. While there is existing research that integrates artificial intelligence in the analysis of various autoimmune diseases, the exploration of dermatomyositis remains relatively underrepresented. In this paper, we present a deep-learning approach tailored for Medical image segmentation. Our proposed method outperforms the current state-of-the-art techniques by an average of 12.26% for U-Net and 12.04% for U-Net++ across the ResNet family of encoders on the dermatomyositis dataset. Furthermore, we probe the importance of optimizing loss function weights and benchmark our methodology on three challenging medical image segmentation tasks
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge