Mei Chen
PISCES: Annotation-free Text-to-Video Post-Training via Optimal Transport-Aligned Rewards
Feb 02, 2026Abstract:Text-to-video (T2V) generation aims to synthesize videos with high visual quality and temporal consistency that are semantically aligned with input text. Reward-based post-training has emerged as a promising direction to improve the quality and semantic alignment of generated videos. However, recent methods either rely on large-scale human preference annotations or operate on misaligned embeddings from pre-trained vision-language models, leading to limited scalability or suboptimal supervision. We present $\texttt{PISCES}$, an annotation-free post-training algorithm that addresses these limitations via a novel Dual Optimal Transport (OT)-aligned Rewards module. To align reward signals with human judgment, $\texttt{PISCES}$ uses OT to bridge text and video embeddings at both distributional and discrete token levels, enabling reward supervision to fulfill two objectives: (i) a Distributional OT-aligned Quality Reward that captures overall visual quality and temporal coherence; and (ii) a Discrete Token-level OT-aligned Semantic Reward that enforces semantic, spatio-temporal correspondence between text and video tokens. To our knowledge, $\texttt{PISCES}$ is the first to improve annotation-free reward supervision in generative post-training through the lens of OT. Experiments on both short- and long-video generation show that $\texttt{PISCES}$ outperforms both annotation-based and annotation-free methods on VBench across Quality and Semantic scores, with human preference studies further validating its effectiveness. We show that the Dual OT-aligned Rewards module is compatible with multiple optimization paradigms, including direct backpropagation and reinforcement learning fine-tuning.
Step-GUI Technical Report
Dec 19, 2025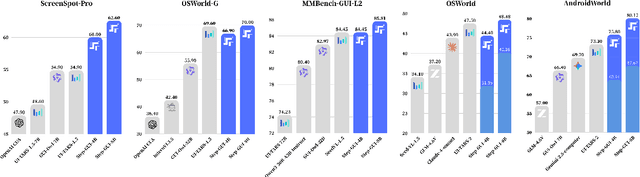
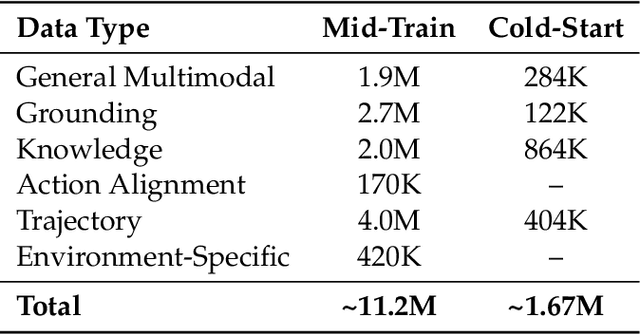
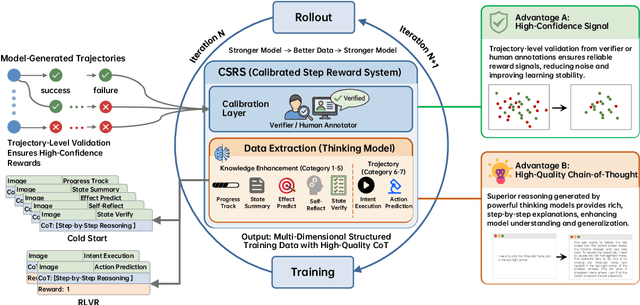

Abstract:Recent advances in multimodal large language models unlock unprecedented opportunities for GUI automation. However, a fundamental challenge remains: how to efficiently acquire high-quality training data while maintaining annotation reliability? We introduce a self-evolving training pipeline powered by the Calibrated Step Reward System, which converts model-generated trajectories into reliable training signals through trajectory-level calibration, achieving >90% annotation accuracy with 10-100x lower cost. Leveraging this pipeline, we introduce Step-GUI, a family of models (4B/8B) that achieves state-of-the-art GUI performance (8B: 80.2% AndroidWorld, 48.5% OSWorld, 62.6% ScreenShot-Pro) while maintaining robust general capabilities. As GUI agent capabilities improve, practical deployment demands standardized interfaces across heterogeneous devices while protecting user privacy. To this end, we propose GUI-MCP, the first Model Context Protocol for GUI automation with hierarchical architecture that combines low-level atomic operations and high-level task delegation to local specialist models, enabling high-privacy execution where sensitive data stays on-device. Finally, to assess whether agents can handle authentic everyday usage, we introduce AndroidDaily, a benchmark grounded in real-world mobile usage patterns with 3146 static actions and 235 end-to-end tasks across high-frequency daily scenarios (8B: static 89.91%, end-to-end 52.50%). Our work advances the development of practical GUI agents and demonstrates strong potential for real-world deployment in everyday digital interactions.
Beta Distribution Learning for Reliable Roadway Crash Risk Assessment
Nov 07, 2025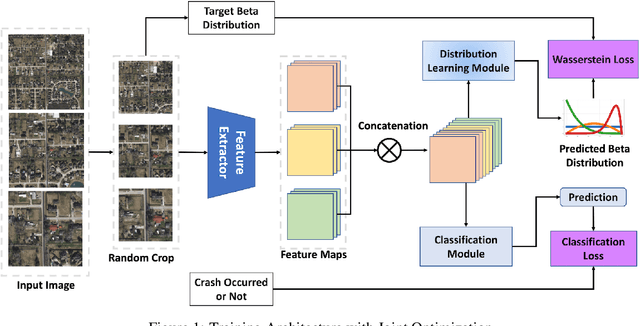
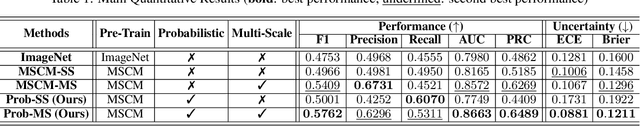
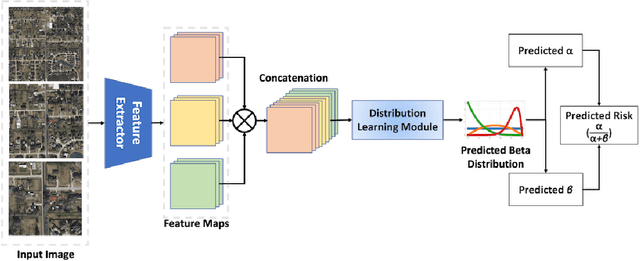
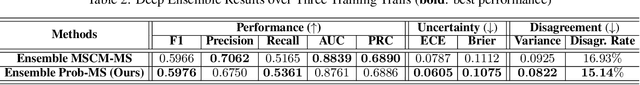
Abstract:Roadway traffic accidents represent a global health crisis, responsible for over a million deaths annually and costing many countries up to 3% of their GDP. Traditional traffic safety studies often examine risk factors in isolation, overlooking the spatial complexity and contextual interactions inherent in the built environment. Furthermore, conventional Neural Network-based risk estimators typically generate point estimates without conveying model uncertainty, limiting their utility in critical decision-making. To address these shortcomings, we introduce a novel geospatial deep learning framework that leverages satellite imagery as a comprehensive spatial input. This approach enables the model to capture the nuanced spatial patterns and embedded environmental risk factors that contribute to fatal crash risks. Rather than producing a single deterministic output, our model estimates a full Beta probability distribution over fatal crash risk, yielding accurate and uncertainty-aware predictions--a critical feature for trustworthy AI in safety-critical applications. Our model outperforms baselines by achieving a 17-23% improvement in recall, a key metric for flagging potential dangers, while delivering superior calibration. By providing reliable and interpretable risk assessments from satellite imagery alone, our method enables safer autonomous navigation and offers a highly scalable tool for urban planners and policymakers to enhance roadway safety equitably and cost-effectively.
DeCafNet: Delegate and Conquer for Efficient Temporal Grounding in Long Videos
May 22, 2025Abstract:Long Video Temporal Grounding (LVTG) aims at identifying specific moments within lengthy videos based on user-provided text queries for effective content retrieval. The approach taken by existing methods of dividing video into clips and processing each clip via a full-scale expert encoder is challenging to scale due to prohibitive computational costs of processing a large number of clips in long videos. To address this issue, we introduce DeCafNet, an approach employing ``delegate-and-conquer'' strategy to achieve computation efficiency without sacrificing grounding performance. DeCafNet introduces a sidekick encoder that performs dense feature extraction over all video clips in a resource-efficient manner, while generating a saliency map to identify the most relevant clips for full processing by the expert encoder. To effectively leverage features from sidekick and expert encoders that exist at different temporal resolutions, we introduce DeCaf-Grounder, which unifies and refines them via query-aware temporal aggregation and multi-scale temporal refinement for accurate grounding. Experiments on two LTVG benchmark datasets demonstrate that DeCafNet reduces computation by up to 47\% while still outperforming existing methods, establishing a new state-of-the-art for LTVG in terms of both efficiency and performance. Our code is available at https://github.com/ZijiaLewisLu/CVPR2025-DeCafNet.
Defense against Prompt Injection Attacks via Mixture of Encodings
Apr 10, 2025Abstract:Large Language Models (LLMs) have emerged as a dominant approach for a wide range of NLP tasks, with their access to external information further enhancing their capabilities. However, this introduces new vulnerabilities, known as prompt injection attacks, where external content embeds malicious instructions that manipulate the LLM's output. Recently, the Base64 defense has been recognized as one of the most effective methods for reducing success rate of prompt injection attacks. Despite its efficacy, this method can degrade LLM performance on certain NLP tasks. To address this challenge, we propose a novel defense mechanism: mixture of encodings, which utilizes multiple character encodings, including Base64. Extensive experimental results show that our method achieves one of the lowest attack success rates under prompt injection attacks, while maintaining high performance across all NLP tasks, outperforming existing character encoding-based defense methods. This underscores the effectiveness of our mixture of encodings strategy for both safety and task performance metrics.
Step-Audio: Unified Understanding and Generation in Intelligent Speech Interaction
Feb 18, 2025Abstract:Real-time speech interaction, serving as a fundamental interface for human-machine collaboration, holds immense potential. However, current open-source models face limitations such as high costs in voice data collection, weakness in dynamic control, and limited intelligence. To address these challenges, this paper introduces Step-Audio, the first production-ready open-source solution. Key contributions include: 1) a 130B-parameter unified speech-text multi-modal model that achieves unified understanding and generation, with the Step-Audio-Chat version open-sourced; 2) a generative speech data engine that establishes an affordable voice cloning framework and produces the open-sourced lightweight Step-Audio-TTS-3B model through distillation; 3) an instruction-driven fine control system enabling dynamic adjustments across dialects, emotions, singing, and RAP; 4) an enhanced cognitive architecture augmented with tool calling and role-playing abilities to manage complex tasks effectively. Based on our new StepEval-Audio-360 evaluation benchmark, Step-Audio achieves state-of-the-art performance in human evaluations, especially in terms of instruction following. On open-source benchmarks like LLaMA Question, shows 9.3% average performance improvement, demonstrating our commitment to advancing the development of open-source multi-modal language technologies. Our code and models are available at https://github.com/stepfun-ai/Step-Audio.
Hummingbird: High Fidelity Image Generation via Multimodal Context Alignment
Feb 07, 2025



Abstract:While diffusion models are powerful in generating high-quality, diverse synthetic data for object-centric tasks, existing methods struggle with scene-aware tasks such as Visual Question Answering (VQA) and Human-Object Interaction (HOI) Reasoning, where it is critical to preserve scene attributes in generated images consistent with a multimodal context, i.e. a reference image with accompanying text guidance query. To address this, we introduce Hummingbird, the first diffusion-based image generator which, given a multimodal context, generates highly diverse images w.r.t. the reference image while ensuring high fidelity by accurately preserving scene attributes, such as object interactions and spatial relationships from the text guidance. Hummingbird employs a novel Multimodal Context Evaluator that simultaneously optimizes our formulated Global Semantic and Fine-grained Consistency Rewards to ensure generated images preserve the scene attributes of reference images in relation to the text guidance while maintaining diversity. As the first model to address the task of maintaining both diversity and fidelity given a multimodal context, we introduce a new benchmark formulation incorporating MME Perception and Bongard HOI datasets. Benchmark experiments show Hummingbird outperforms all existing methods by achieving superior fidelity while maintaining diversity, validating Hummingbird's potential as a robust multimodal context-aligned image generator in complex visual tasks.
PediaBench: A Comprehensive Chinese Pediatric Dataset for Benchmarking Large Language Models
Dec 09, 2024



Abstract:The emergence of Large Language Models (LLMs) in the medical domain has stressed a compelling need for standard datasets to evaluate their question-answering (QA) performance. Although there have been several benchmark datasets for medical QA, they either cover common knowledge across different departments or are specific to another department rather than pediatrics. Moreover, some of them are limited to objective questions and do not measure the generation capacity of LLMs. Therefore, they cannot comprehensively assess the QA ability of LLMs in pediatrics. To fill this gap, we construct PediaBench, the first Chinese pediatric dataset for LLM evaluation. Specifically, it contains 4,565 objective questions and 1,632 subjective questions spanning 12 pediatric disease groups. It adopts an integrated scoring criterion based on different difficulty levels to thoroughly assess the proficiency of an LLM in instruction following, knowledge understanding, clinical case analysis, etc. Finally, we validate the effectiveness of PediaBench with extensive experiments on 20 open-source and commercial LLMs. Through an in-depth analysis of experimental results, we offer insights into the ability of LLMs to answer pediatric questions in the Chinese context, highlighting their limitations for further improvements. Our code and data are published at https://github.com/ACMISLab/PediaBench.
SplatFlow: Self-Supervised Dynamic Gaussian Splatting in Neural Motion Flow Field for Autonomous Driving
Nov 23, 2024



Abstract:Most existing Dynamic Gaussian Splatting methods for complex dynamic urban scenarios rely on accurate object-level supervision from expensive manual labeling, limiting their scalability in real-world applications. In this paper, we introduce SplatFlow, a Self-Supervised Dynamic Gaussian Splatting within Neural Motion Flow Fields (NMFF) to learn 4D space-time representations without requiring tracked 3D bounding boxes, enabling accurate dynamic scene reconstruction and novel view RGB, depth and flow synthesis. SplatFlow designs a unified framework to seamlessly integrate time-dependent 4D Gaussian representation within NMFF, where NMFF is a set of implicit functions to model temporal motions of both LiDAR points and Gaussians as continuous motion flow fields. Leveraging NMFF, SplatFlow effectively decomposes static background and dynamic objects, representing them with 3D and 4D Gaussian primitives, respectively. NMFF also models the status correspondences of each 4D Gaussian across time, which aggregates temporal features to enhance cross-view consistency of dynamic components. SplatFlow further improves dynamic scene identification by distilling features from 2D foundational models into 4D space-time representation. Comprehensive evaluations conducted on the Waymo Open Dataset and KITTI Dataset validate SplatFlow's state-of-the-art (SOTA) performance for both image reconstruction and novel view synthesis in dynamic urban scenarios.
Cell as Point: One-Stage Framework for Efficient Cell Tracking
Nov 22, 2024



Abstract:Cellular activities are dynamic and intricate, playing a crucial role in advancing diagnostic and therapeutic techniques, yet they often require substantial resources for accurate tracking. Despite recent progress, the conventional multi-stage cell tracking approaches not only heavily rely on detection or segmentation results as a prerequisite for the tracking stage, demanding plenty of refined segmentation masks, but are also deteriorated by imbalanced and long sequence data, leading to under-learning in training and missing cells in inference procedures. To alleviate the above issues, this paper proposes the novel end-to-end CAP framework, which leverages the idea of regarding Cell as Point to achieve efficient and stable cell tracking in one stage. CAP abandons detection or segmentation stages and simplifies the process by exploiting the correlation among the trajectories of cell points to track cells jointly, thus reducing the label demand and complexity of the pipeline. With cell point trajectory and visibility to represent cell locations and lineage relationships, CAP leverages the key innovations of adaptive event-guided (AEG) sampling for addressing data imbalance in cell division events and the rolling-as-window (RAW) inference method to ensure continuous tracking of new cells in the long term. Eliminating the need for a prerequisite detection or segmentation stage, CAP demonstrates strong cell tracking performance while also being 10 to 55 times more efficient than existing methods. The code and models will be released.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge